3E6I
 
 | |
3VEX
 
 | | Crystal structure of the O-carbamoyltransferase TobZ H14N variant in complex with carbamoyl adenylate intermediate | | Descriptor: | 1,2-ETHANEDIOL, 5'-O-[(S)-(carbamoyloxy)(hydroxy)phosphoryl]adenosine, FE (II) ION, ... | | Authors: | Parthier, C, Stubbs, M.T, Goerlich, S, Jaenecke, F. | | Deposit date: | 2012-01-09 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The O-Carbamoyltransferase TobZ Catalyzes an Ancient Enzymatic Reaction.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
3MV5
 
 | | Crystal structure of Akt-1-inhibitor complexes | | Descriptor: | (3R)-1-(5-methyl-7H-pyrrolo[2,3-d]pyrimidin-4-yl)pyrrolidin-3-amine, GSK3-beta peptide, MANGANESE (II) ION, ... | | Authors: | Pandit, J. | | Deposit date: | 2010-05-03 | | Release date: | 2010-06-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Design of selective, ATP-competitive inhibitors of Akt.
J.Med.Chem., 53, 2010
|
|
3EDG
 
 | |
3VFE
 
 | | Virtual Screening and X-Ray Crystallography for Human Kallikrein 6 Inhibitors with an Amidinothiophene P1 Group | | Descriptor: | 4-{[(3R)-3-{[(7-methoxynaphthalen-2-yl)sulfonyl](thiophen-3-ylmethyl)amino}-2-oxopyrrolidin-1-yl]methyl}thiophene-2-carboximidamide, Kallikrein-6 | | Authors: | Chen, X, Zhang, Y, Xia, T, Wang, R. | | Deposit date: | 2012-01-09 | | Release date: | 2012-11-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Virtual Screening and X-ray Crystallography for Human Kallikrein 6 Inhibitors with an Amidinothiophene P1 Group.
Acs Med.Chem.Lett., 3, 2012
|
|
3EDN
 
 | | Crystal structure of the Bacillus anthracis phenazine biosynthesis protein, PhzF family | | Descriptor: | MAGNESIUM ION, Phenazine biosynthesis protein, PhzF family, ... | | Authors: | Anderson, S.M, Brunzelle, J.S, Onopriyenko, O, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2008-09-03 | | Release date: | 2008-10-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the Bacillus anthracis phenazine biosynthesis protein, PhzF family
To be Published
|
|
3VFV
 
 | | crystal structure of HLA B*3508 LPEP-P9Ala, peptide mutant P9-ala | | Descriptor: | Beta-2-microglobulin, LPEP peptide from EBV, P9A, ... | | Authors: | Liu, Y.C, Rossjohn, J, Gras, S. | | Deposit date: | 2012-01-10 | | Release date: | 2012-02-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The Energetic Basis Underpinning T-cell Receptor Recognition of a Super-bulged Peptide Bound to a Major Histocompatibility Complex Class I Molecule.
J.Biol.Chem., 287, 2012
|
|
3E7K
 
 | |
3VGE
 
 | | Crystal structure of glycosyltrehalose trehalohydrolase (D252S) | | Descriptor: | CITRATE ANION, GLYCEROL, Malto-oligosyltrehalose trehalohydrolase | | Authors: | Okazaki, N, Tamada, T, Feese, M.D, Kato, M, Miura, Y, Komeda, T, Kobayashi, K, Kondo, K, Kuroki, R. | | Deposit date: | 2011-08-09 | | Release date: | 2012-06-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Substrate recognition mechanism of a glycosyltrehalose trehalohydrolase from Sulfolobus solfataricus KM1.
Protein Sci., 21, 2012
|
|
3MXP
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS T62A at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Caro, J.A, Schlessman, J.L, Garcia-Moreno, E.B, Heroux, A. | | Deposit date: | 2010-05-07 | | Release date: | 2011-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Cavities determine the pressure unfolding of proteins.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3E7W
 
 | | Crystal structure of DLTA: Implications for the reaction mechanism of non-ribosomal peptide synthetase (NRPS) adenylation domains | | Descriptor: | ADENOSINE MONOPHOSPHATE, D-alanine--poly(phosphoribitol) ligase subunit 1, PHOSPHATE ION | | Authors: | Yonus, H, Neumann, P, Zimmermann, S, May, J.J, Marahiel, M.A, Stubbs, M.T. | | Deposit date: | 2008-08-19 | | Release date: | 2008-09-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structure of DltA. Implications for the reaction mechanism of non-ribosomal peptide synthetase adenylation domains
J.Biol.Chem., 283, 2008
|
|
3MV9
 
 | | Crystal Structure of the TK3-Gln55Ala TCR in complex with HLA-B*3501/HPVG | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-35 alpha chain, ... | | Authors: | Gras, S, Chen, Z, Miles, J.J, Liu, Y.C, Bell, M.J, Sullivan, L.C, Kjer-Nielsen, L, Brennan, R.M, Burrows, J.M, Neller, M.A, Khanna, R, Purcell, A.W, Brooks, A.G, McCluskey, J, Rossjohn, J, Burrows, S.R. | | Deposit date: | 2010-05-03 | | Release date: | 2010-06-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Allelic polymorphism in the T cell receptor and its impact on immune responses
J.Exp.Med., 207, 2010
|
|
3VHD
 
 | | Hsp90 alpha N-terminal domain in complex with a macrocyclic inhibitor, CH5164840 | | Descriptor: | 4-amino-18,20-dimethyl-7-thia-3,5,11,15-tetraazatricyclo[15.3.1.1(2,6)]docosa-1(20),2,4,6(22),17(21),18-hexaene-10,16-dione, Heat shock protein HSP 90-alpha | | Authors: | Fukami, T.A, Ono, N. | | Deposit date: | 2011-08-24 | | Release date: | 2012-07-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Design and synthesis of novel macrocyclic 2-amino-6-arylpyrimidine Hsp90 inhibitors
Bioorg.Med.Chem.Lett., 22, 2012
|
|
3MVJ
 
 | | Human cyclic AMP-dependent protein kinase PKA inhibitor complex | | Descriptor: | (3R)-1-(5-methyl-7H-pyrrolo[2,3-d]pyrimidin-4-yl)pyrrolidin-3-amine, cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha | | Authors: | Pandit, J, Vajdos, F. | | Deposit date: | 2010-05-04 | | Release date: | 2010-06-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Design of selective, ATP-competitive inhibitors of Akt.
J.Med.Chem., 53, 2010
|
|
3MYD
 
 | |
3EMQ
 
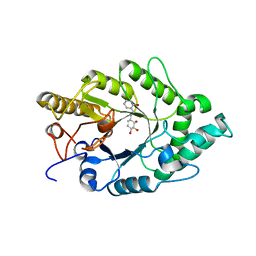 | | Crystal structure of xilanase XynB from Paenibacillus barcelonensis complexed with an inhibitor | | Descriptor: | (1S,2S,3R,6R)-6-[(2-hydroxybenzyl)amino]cyclohex-4-ene-1,2,3-triol, Endo-1,4-beta-xylanase | | Authors: | Sanz-Aparicio, J, Isorna, P. | | Deposit date: | 2008-09-25 | | Release date: | 2009-09-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Structural insights into the specificity of Xyn10B from Paenibacillus barcinonensis and its improved stability by forced protein evolution.
J.Biol.Chem., 285, 2010
|
|
3E98
 
 | |
3VJF
 
 | | Crystal structure of de novo 4-helix bundle protein WA20 | | Descriptor: | POTASSIUM ION, WA20 | | Authors: | Arai, R, Kimura, A, Kobayashi, N, Matsuo, K, Sato, T, Wang, A.F, Platt, J.M, Bradley, L.H, Hecht, M.H. | | Deposit date: | 2011-10-18 | | Release date: | 2012-03-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Domain-swapped dimeric structure of a stable and functional de novo four-helix bundle protein, WA20
J.Phys.Chem.B, 116, 2012
|
|
3E9N
 
 | | Crystal structure of a putative short-chain dehydrogenase/reductase from Corynebacterium glutamicum | | Descriptor: | PUTATIVE SHORT-CHAIN DEHYDROGENASE/REDUCTASE | | Authors: | Bonanno, J.B, Gilmore, M, Bain, K.T, Hu, S, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-08-22 | | Release date: | 2008-09-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of a putative short-chain dehydrogenase/reductase from Corynebacterium glutamicum
To be Published
|
|
3EN5
 
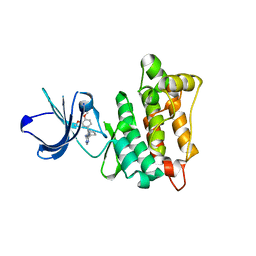 | | Targeted polypharmacology: crystal structure of the c-Src kinase domain in complex with PP494, a multitargeted kinase inhibitor | | Descriptor: | 1-cyclobutyl-3-(3,4-dimethoxyphenyl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Blair, J.A, Apsel, B, Knight, Z.A, Shokat, K.M. | | Deposit date: | 2008-09-25 | | Release date: | 2008-10-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Targeted polypharmacology: discovery of dual inhibitors of tyrosine and phosphoinositide kinases.
Nat.Chem.Biol., 4, 2008
|
|
3MZ5
 
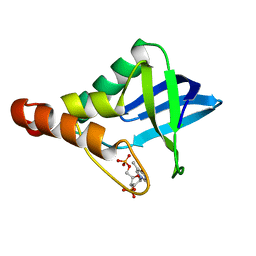 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS L103A at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Caro, J.A, Schlessman, J.L, Garcia-Moreno, E.B, Heroux, A. | | Deposit date: | 2010-05-11 | | Release date: | 2011-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Cavities determine the pressure unfolding of proteins.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3VL3
 
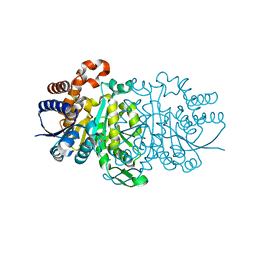 | |
3VLX
 
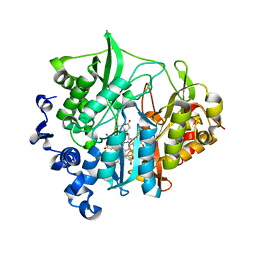 | | Assimilatory nitrite reductase (Nii3) - N226K mutant - ligand free form from tobacco leaf | | Descriptor: | CHLORIDE ION, IRON/SULFUR CLUSTER, Nitrite reductase, ... | | Authors: | Nakano, S, Takahashi, M, Sakamoto, A, Morikawa, H, Katayanagi, K. | | Deposit date: | 2011-12-05 | | Release date: | 2012-09-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | X-ray crystal structure of a mutant assimilatory nitrite reductase that shows sulfite reductase-like activity
Chem.Biodivers., 9, 2012
|
|
3EOP
 
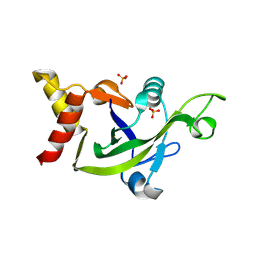 | | Crystal Structure of the DUF55 domain of human thymocyte nuclear protein 1 | | Descriptor: | SULFATE ION, Thymocyte nuclear protein 1 | | Authors: | Yu, F, Song, A, Xu, C, Sun, L, Li, L, Tang, L, Hu, H, He, J. | | Deposit date: | 2008-09-29 | | Release date: | 2009-09-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Determining the DUF55-domain structure of human thymocyte nuclear protein 1 from crystals partially twinned by tetartohedry
Acta Crystallogr.,Sect.D, 65, 2009
|
|
3MZO
 
 | |
