1HAA
 
 | | A beta-Hairpin Structure in a 13-mer Peptide that Binds a-Bungarotoxin with High Affinity and Neutralizes its Toxicity | | Descriptor: | ALPHA-BUNGAROTOXIN, PEPTIDE | | Authors: | Scherf, T, Kasher, R, Balass, M, Fridkin, M, Fuchs, S, Katchalski-Katzir, E. | | Deposit date: | 2001-04-05 | | Release date: | 2001-05-25 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | A Beta-Hairpin Structure in a 13-mer Peptide that Binds Alpha-Bungarotoxin with High Affinity and Neutralizes its Toxicity
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
2KMG
 
 | | The structure of the KlcA and ArdB proteins show a novel fold and antirestriction activity against Type I DNA restriction systems in vivo but not in vitro | | Descriptor: | KlcA | | Authors: | Serfiotis-Mitsa, D, Herbert, A.P, Roberts, G.A, Soares, D.C, White, J.H, Blakely, G.W, Uhrin, D, Dryden, D.T.F. | | Deposit date: | 2009-07-28 | | Release date: | 2009-12-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The structure of the KlcA and ArdB proteins reveals a novel fold and antirestriction activity against Type I DNA restriction systems in vivo but not in vitro
Nucleic Acids Res., 38, 2010
|
|
2GQN
 
 | |
1HAJ
 
 | | A beta-Hairpin Structure in a 13-mer Peptide that Binds a-Bungarotoxin with High Affinity and Neutralizes its Toxicity | | Descriptor: | ALPHA-BUNGAROTOXIN, PEPTIDE | | Authors: | Scherf, T, Kasher, R, Balass, M, Fridkin, M, Fuchs, S, Katchalski-Katzir, E. | | Deposit date: | 2001-04-05 | | Release date: | 2001-05-25 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | A Beta-Hairpin Structure in a 13-mer Peptide that Binds Alpha-Bungarotoxin with High Affinity and Neutralizes its Toxicity
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
2OP4
 
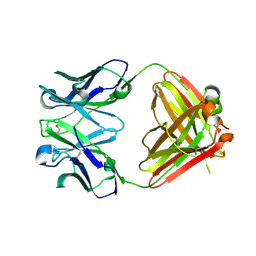 | | Crystal Structure of Quorum-Quenching Antibody 1G9 | | Descriptor: | 1,2-ETHANEDIOL, Murine Antibody Fab RS2-1G9 IGG1 Heavy Chain, Murine Antibody Fab RS2-1G9 Lambda Light Chain | | Authors: | Kirchdoerfer, R.N, Debler, E.W, Wilson, I.A. | | Deposit date: | 2007-01-26 | | Release date: | 2007-05-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal Structures of a Quorum-quenching Antibody.
J.Mol.Biol., 368, 2007
|
|
1GHC
 
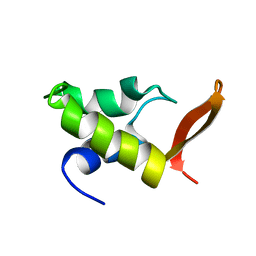 | | HOMO-AND HETERONUCLEAR TWO-DIMENSIONAL NMR STUDIES OF THE GLOBULAR DOMAIN OF HISTONE H1: FULL ASSIGNMENT, TERTIARY STRUCTURE, AND COMPARISON WITH THE GLOBULAR DOMAIN OF HISTONE H5 | | Descriptor: | GH1 | | Authors: | Cerf, C, Lippens, G, Ramakrishnan, V, Muyldermans, S, Segers, A, Wyns, L, Wodak, S.J, Hallenga, K. | | Deposit date: | 1994-05-16 | | Release date: | 1994-08-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Homo- and heteronuclear two-dimensional NMR studies of the globular domain of histone H1: full assignment, tertiary structure, and comparison with the globular domain of histone H5.
Biochemistry, 33, 1994
|
|
1K38
 
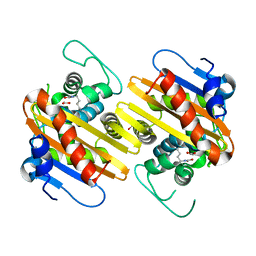 | | CRYSTAL STRUCTURE OF THE CLASS D BETA-LACTAMASE OXA-2 | | Descriptor: | Beta-lactamase OXA-2, FORMIC ACID | | Authors: | Kerff, F, Fonze, E, Bouillenne, F, Frere, J.M, Charlier, P. | | Deposit date: | 2001-10-02 | | Release date: | 2003-06-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | CRYSTAL STRUCTURE OF THE CLASS D BETA-LACTAMASE OXA-2
To be Published
|
|
1K6R
 
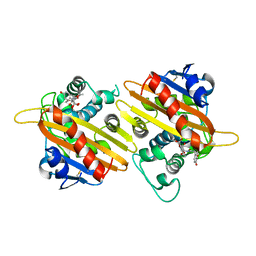 | | STRUCTURE OF THE CLASS D BETA-LACTAMASE OXA-10 IN COMPLEX WITH MOXALACTAM | | Descriptor: | (2R)-2-((R)-CARBOXY{[CARBOXY(4-HYDROXYPHENYL)ACETYL]AMINO}METHOXYMETHYL)-5-METHYLENE-5,6-DIHYDRO-2H-1,3-OXAZINE-4-CARBO XYLIC ACID, Beta-lactamase PSE-2 | | Authors: | Kerff, F, Fonze, E, Sauvage, E, Frere, J.M, Charlier, P. | | Deposit date: | 2001-10-17 | | Release date: | 2003-06-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | CRYSTAL STRUCTURE OF CLASS D BETA-LACTAMASE OXA-10 IN COMPLEX WITH DIFFERENT SUBSTRATES AND ONE INHIBITOR.
To be Published
|
|
1K4E
 
 | | CRYSTAL STRUCTURE OF THE CLASS D BETA-LACTAMASES OXA-10 DETERMINED BY MAD PHASING WITH SELENOMETHIONINE | | Descriptor: | Beta-lactamase PSE-2, SULFATE ION | | Authors: | Kerff, F, Fonze, E, Bouillene, F, Frere, J.M, Charlier, P. | | Deposit date: | 2001-10-08 | | Release date: | 2001-10-31 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | STRUCTURE OF CLASS D BETA-LACTAMASE OXA-2
To be Published
|
|
1K6S
 
 | | STRUCTURE OF THE CLASS D BETA-LACTAMASE OXA-10 IN COMPLEX WITH A PHENYLBORONIC ACID | | Descriptor: | 4-IODO-ACETAMIDO PHENYLBORONIC ACID, Beta-lactamase PSE-2, CALCIUM ION, ... | | Authors: | Kerff, F, Fonze, E, Sauvage, E, Frere, J.M, Charlier, P. | | Deposit date: | 2001-10-17 | | Release date: | 2003-06-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | CRYSTAL STRUCTURE OF CLASS D BETA-LACTAMASE OXA-10 IN COMPLEX WITH DIFFERENT SUBSTRATES AND ONE INHIBITOR.
To be Published
|
|
1KXO
 
 | | ENGINEERED LIPOCALIN DIGA16 : APO-FORM | | Descriptor: | DigA16 | | Authors: | Korndoerfer, I.P, Skerra, A. | | Deposit date: | 2002-02-01 | | Release date: | 2003-06-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural mechanism of specific ligand recognition by a lipocalin tailored for the complexation of digoxigenin.
J.Mol.Biol., 330, 2003
|
|
1LNM
 
 | |
1LKE
 
 | |
1K4F
 
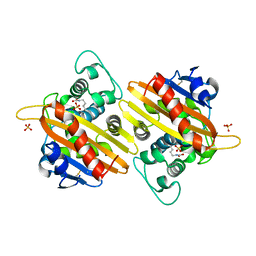 | | CRYSTAL STRUCTURE OF THE CLASS D BETA-LACTAMASE OXA-10 AT 1.6 A RESOLUTION | | Descriptor: | Beta-lactamase PSE-2, SULFATE ION | | Authors: | Kerff, F, Fonze, E, Bouillene, F, Frere, J.M, Charlier, P. | | Deposit date: | 2001-10-08 | | Release date: | 2001-10-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the class D beta-lactamase OXA-2
To be Published
|
|
1KAO
 
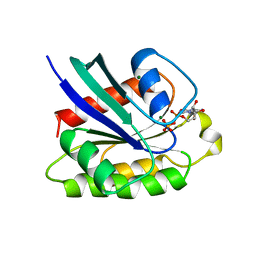 | | CRYSTAL STRUCTURE OF THE SMALL G PROTEIN RAP2A WITH GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, RAP2A | | Authors: | Cherfils, J, Menetrey, J, Le Bras, G. | | Deposit date: | 1997-08-01 | | Release date: | 1997-12-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of the small G protein Rap2A in complex with its substrate GTP, with GDP and with GTPgammaS.
EMBO J., 16, 1997
|
|
1SVD
 
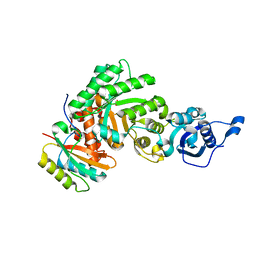 | | The structure of Halothiobacillus neapolitanus RuBisCo | | Descriptor: | GLYCEROL, Ribulose bisphosphate carboxylase small chain, SULFATE ION, ... | | Authors: | Kerfeld, C.A, Sawaya, M.R, Pashkov, I, Cannon, G, Williams, E, Tran, K, Yeates, T.O. | | Deposit date: | 2004-03-29 | | Release date: | 2005-04-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of Halothiobacillus neapolitanus RuBisCo
To be Published
|
|
3BWM
 
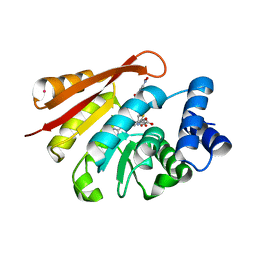 | | Crystal Structure of Human Catechol O-Methyltransferase with bound SAM and DNC | | Descriptor: | 3,5-DINITROCATECHOL, Catechol O-methyltransferase, MAGNESIUM ION, ... | | Authors: | Rutherford, K, Le Trong, I, Stenkamp, R.E, Parson, W.W. | | Deposit date: | 2008-01-09 | | Release date: | 2008-06-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal structures of human 108V and 108M catechol O-methyltransferase.
J.Mol.Biol., 380, 2008
|
|
3BWY
 
 | | Crystal Structure of Human 108M Catechol O-methyltransferase bound with S-adenosylmethionine and inhibitor dinitrocatechol | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3,5-DINITROCATECHOL, COMT protein, ... | | Authors: | Rutherford, K, Le Trong, I, Stenkamp, R.E, Parson, W.W. | | Deposit date: | 2008-01-10 | | Release date: | 2008-06-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structures of human 108V and 108M catechol O-methyltransferase.
J.Mol.Biol., 380, 2008
|
|
1MZ4
 
 | | Crystal Structure of Cytochrome c550 from Thermosynechococcus elongatus | | Descriptor: | BICARBONATE ION, GLYCEROL, HEME C, ... | | Authors: | Kerfeld, C.A, Sawaya, M.R, Bottin, H, Tran, K.T, Sugiura, M, Kirilovsky, D, Krogmann, D, Yeates, T.O, Boussac, A. | | Deposit date: | 2002-10-05 | | Release date: | 2003-09-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and EPR characterization of the soluble form of cytochrome c-550 and of the psbV2 gene product from the cyanobacterium Thermosynechococcus elongatus.
Plant Cell.Physiol., 44, 2003
|
|
3QG6
 
 | | Structural Basis for Ligand Recognition and Discrimination of a Quorum Quenching Antibody | | Descriptor: | AP4-24H11 Heavy Chain, AP4-24H11 Light Chain, Agr autoinducing peptide, ... | | Authors: | Kirchdoerfer, R.N, Janda, J.D, Kaufmann, G.F, Wilson, I.A. | | Deposit date: | 2011-01-24 | | Release date: | 2011-03-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for Ligand Recognition and Discrimination of a Quorum-quenching Antibody.
J.Biol.Chem., 286, 2011
|
|
3TWI
 
 | | Variable Lymphocyte Receptor Recognition of the Immunodominant Glycoprotein of Bacillus anthracis Spores | | Descriptor: | BclA protein, GLYCEROL, Variable lymphocyte receptor B | | Authors: | Kirchdoerfer, R.N, Herrin, B.R, Han, B.W, Turnbough Jr, C.L, Cooper, M.D, Wilson, I.A. | | Deposit date: | 2011-09-21 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Variable Lymphocyte Receptor Recognition of the Immunodominant Glycoprotein of Bacillus anthracis Spores.
Structure, 20, 2012
|
|
3TYJ
 
 | | Bacillus collagen-like protein of anthracis P159S mutant | | Descriptor: | BclA protein | | Authors: | Kirchdoerfer, R.N, Herrin, B.R, Han, B.W, Turnbough Jr, C.L, Cooper, M.D, Wilson, I.A. | | Deposit date: | 2011-09-26 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Variable Lymphocyte Receptor Recognition of the Immunodominant Glycoprotein of Bacillus anthracis Spores.
Structure, 20, 2012
|
|
4ZTG
 
 | |
4ZTI
 
 | |
4ZTA
 
 | |
