2AFV
 
 | | The Crystal Structure of Putative Precorrin Isomerase CbiC in Cobalamin Biosynthesis | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, CHLORIDE ION, HEXAETHYLENE GLYCOL, ... | | Authors: | Xue, Y, Wei, Z. | | Deposit date: | 2005-07-26 | | Release date: | 2006-04-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structure of putative precorrin isomerase CbiC in cobalamin biosynthesis
J.Struct.Biol., 153, 2006
|
|
4K2I
 
 | | Crystal structure of ntda from bacillus subtilis with bound cofactor pmp | | Descriptor: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, ACETATE ION, ... | | Authors: | Van Straaten, K.E, Palmer, D.R.J, Sanders, D.A.R. | | Deposit date: | 2013-04-09 | | Release date: | 2013-10-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.225 Å) | | Cite: | The Structure of NtdA, a Sugar Aminotransferase Involved in the Kanosamine Biosynthetic Pathway in Bacillus subtilis, Reveals a New Subclass of Aminotransferases.
J.Biol.Chem., 288, 2013
|
|
3PSB
 
 | | Furo[2,3-c]pyridine-based Indanone Oximes as Potent and Selective B-Raf Inhibitors | | Descriptor: | B-RAF PROTO-ONCOGENE SERINE/THREONINE-PROTEIN KINASE, ethyl 3-{[1-(hydroxyamino)-2H-inden-5-yl]amino}thieno[2,3-c]pyridine-2-carboxylate | | Authors: | Morales, T, Vigers, G.P.A, Brandhuber, B.J. | | Deposit date: | 2010-12-01 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The Discovery of furo[2,3-c]pyridine-based indanone oximes as potent and selective B-Raf inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3PSW
 
 | | Structure of E97Q mutant of TIM from Plasmodium falciparum | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Triosephosphate isomerase | | Authors: | Samanta, M, Murthy, M.R.N, Balaram, H, Balaram, P. | | Deposit date: | 2010-12-02 | | Release date: | 2011-10-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Revisiting the mechanism of the triosephosphate isomerase reaction: the role of the fully conserved glutamic acid 97 residue
Chembiochem, 12, 2011
|
|
3ABQ
 
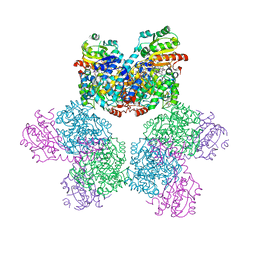 | |
6FF7
 
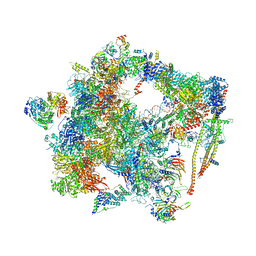 | | human Bact spliceosome core structure | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, ADENOSINE-5'-DIPHOSPHATE, BUD13 homolog, ... | | Authors: | Haselbach, D, Komarov, I, Agafonov, D, Hartmuth, K, Graf, B, Kastner, B, Luehrmann, R, Stark, H. | | Deposit date: | 2018-01-03 | | Release date: | 2019-03-13 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structure and Conformational Dynamics of the Human Spliceosomal BactComplex.
Cell, 172, 2018
|
|
1WXV
 
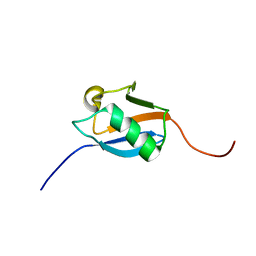 | | Solution structure of the ubiquitin domain of BCL-2 binding athanogene-1 | | Descriptor: | BAG-family molecular chaperone regulator-1 | | Authors: | Niraula, T.N, Muto, Y, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-02-02 | | Release date: | 2005-08-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the ubiquitin domain of BCL-2 binding athanogene-1
To be Published
|
|
5ACQ
 
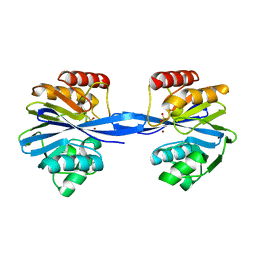 | | W228A-Investigation of the impact from residues W228 and Y233 in the metallo-beta-lactamase GIM-1 | | Descriptor: | BETA-LACTAMASE, ZINC ION | | Authors: | Skagseth, S, Carlsen, T.J, Bjerga, G.E.K, Spencer, J, Samuelsen, O, Leiros, H.-K.S. | | Deposit date: | 2015-08-17 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Role of Residues W228 and Y233 in the Structure and Activity of Metallo-Beta-Lactamase Gim-1.
Antimicrob.Agents Chemother., 60, 2015
|
|
6KPQ
 
 | | Crystal structure of Zika NS2B-NS3 protease with compound 8 | | Descriptor: | 1-[(10~{R},17~{S},20~{S})-17,20-bis(4-azanylbutyl)-4,9,16,19,22-pentakis(oxidanylidene)-3,8,15,18,21-pentazabicyclo[22.3.1]octacosa-1(27),24(28),25-trien-10-yl]guanidine, NS3 protease, Serine protease subunit NS2B | | Authors: | Quek, J.P. | | Deposit date: | 2019-08-15 | | Release date: | 2020-06-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structure-Based Macrocyclization of Substrate Analogue NS2B-NS3 Protease Inhibitors of Zika, West Nile and Dengue viruses.
Chemmedchem, 15, 2020
|
|
2V8O
 
 | | Structure of the Murray Valley encephalitis virus RNA helicase to 1. 9A resolution | | Descriptor: | FLAVIVIRIN PROTEASE NS3 | | Authors: | Mancini, E.J, Assenberg, R, Verma, A, Walter, T.S, Tuma, R, Grimes, J.M, Owens, R.J, Stuart, D.I. | | Deposit date: | 2007-08-09 | | Release date: | 2007-08-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the Murray Valley Encephalitis Virus RNA Helicase at 1.9 A Resolution.
Protein Sci., 16, 2007
|
|
5QIK
 
 | | TGF-BETA RECEPTOR TYPE 1 KINASE DOMAIN (T204D) IN COMPLEX WITH N-{4-[3-(6-fluoropyridin-3-yl)-4-oxo-4,5,6,7-tetrahydro-1H-pyrrolo[3,2-c]pyridin-2-yl]pyridin-2-yl}acetamide | | Descriptor: | GLYCEROL, N-{4-[3-(6-fluoropyridin-3-yl)-4-oxo-4,5,6,7-tetrahydro-1H-pyrrolo[3,2-c]pyridin-2-yl]pyridin-2-yl}acetamide, TGF-beta receptor type-1 | | Authors: | Sheriff, S. | | Deposit date: | 2018-08-05 | | Release date: | 2018-10-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Discovery of 4-Azaindole Inhibitors of TGF beta RI as Immuno-oncology Agents.
ACS Med Chem Lett, 9, 2018
|
|
3Q33
 
 | | Structure of the Rtt109-AcCoA/Vps75 Complex and Implications for Chaperone-Mediated Histone Acetylation | | Descriptor: | 1,2-ETHANEDIOL, ACETYL COENZYME *A, HISTONE H3, ... | | Authors: | Tang, Y, Yuan, H, Meeth, K, Marmorstein, R. | | Deposit date: | 2010-12-21 | | Release date: | 2011-02-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the Rtt109-AcCoA/Vps75 Complex and Implications for Chaperone-Mediated Histone Acetylation.
Structure, 19, 2011
|
|
4D9E
 
 | | D-Cysteine desulfhydrase from Salmonella typhimurium complexed with L-cycloserine (LCS) | | Descriptor: | BENZAMIDINE, D-Cysteine desulfhydrase, [5-hydroxy-6-methyl-4-({[(4E)-3-oxo-1,2-oxazolidin-4-ylidene]amino}methyl)pyridin-3-yl]methyl dihydrogen phosphate | | Authors: | Bharath, S.R, Shveta, B, Rajesh, K.H, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2012-01-11 | | Release date: | 2012-05-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Structural and Mutational Studies on Substrate Specificity and Catalysis of Salmonella typhimurium D-Cysteine Desulfhydrase.
Plos One, 7, 2012
|
|
5ACY
 
 | | N-TERMINAL BROMODOMAIN OF HUMAN BRD4 WITH 1-(2R,4S)-2-methyl-4-(phenylamino)-6-4-(piperidin-1-ylmethyl)phenyl-1,2,3,4- tetrahydroquinolin-1-yl-ethan-1-one | | Descriptor: | 1,2-ETHANEDIOL, 1-[(2S,4R)-2-methyl-4-(phenylamino)-6-[4-(piperidin-1-ylmethyl)phenyl]-3,4-dihydroquinolin-1(2H)-yl]ethanone, BROMODOMAIN-CONTAINING PROTEIN 4 | | Authors: | Chung, C. | | Deposit date: | 2015-08-18 | | Release date: | 2015-10-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Autism-Like Syndrome is Induced by Pharmacological Suppression of Bet Proteins in Young Mice.
J.Exp.Med., 212, 2015
|
|
3M81
 
 | |
1QDQ
 
 | | X-RAY CRYSTAL STRUCTURE OF BOVINE CATHEPSIN B-CA074 COMPLEX | | Descriptor: | CATHEPSIN B, [PROPYLAMINO-3-HYDROXY-BUTAN-1,4-DIONYL]-ISOLEUCYL-PROLINE | | Authors: | Yamamoto, A. | | Deposit date: | 1999-07-10 | | Release date: | 2000-07-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Substrate specificity of bovine cathepsin B and its inhibition by CA074, based on crystal structure refinement of the complex.
J.Biochem.(Tokyo), 127, 2000
|
|
2Q9J
 
 | | Crystal structure of the C217S mutant of diaminopimelate epimerase | | Descriptor: | 1,2-ETHANEDIOL, Diaminopimelate epimerase, SULFATE ION | | Authors: | Pillai, B, Cherney, M, Diaper, C.M, Sutherland, A, Blanchard, J.S, Vederas, J.C. | | Deposit date: | 2007-06-12 | | Release date: | 2007-10-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dynamics of catalysis revealed from the crystal structures of mutants of diaminopimelate epimerase.
Biochem.Biophys.Res.Commun., 363, 2007
|
|
1X57
 
 | | Solution structures of the HTH domain of human EDF-1 protein | | Descriptor: | Endothelial differentiation-related factor 1 | | Authors: | Nameki, N, Sato, M, Tochio, N, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-15 | | Release date: | 2005-11-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the HTH domain of human EDF-1 protein
To be Published
|
|
4D08
 
 | | PDE2a catalytic domain in complex with a brain penetrant inhibitor | | Descriptor: | 1-(5-butoxypyridin-3-yl)-4-methyl-8-(morpholin-4-ylmethyl)[1,2,4]triazolo[4,3-a]quinoxaline, CGMP-DEPENDENT 3', 5'-CYCLIC PHOSPHODIESTERASE, ... | | Authors: | Buijnsters, P, Andres, J.I, DeAngelis, M, Langlois, X, Rombouts, F, Sanderson, W, Tresadern, G, Trabanco, A, VanHoof, G, VanRoosbroeck, Y. | | Deposit date: | 2014-04-24 | | Release date: | 2014-08-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Based Design of a Potent, Selective, and Brain Penetrating Pde2 Inhibitor with Demonstrated Target Engagement.
Acs Med.Chem.Lett., 5, 2014
|
|
3VES
 
 | | Crystal structure of the O-carbamoyltransferase TobZ in complex with AMPCPP and carbamoyl phosphate | | Descriptor: | 1,2-ETHANEDIOL, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, FE (II) ION, ... | | Authors: | Parthier, C, Stubbs, M.T, Goerlich, S, Jaenecke, F. | | Deposit date: | 2012-01-09 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | The O-Carbamoyltransferase TobZ Catalyzes an Ancient Enzymatic Reaction.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
3AHG
 
 | | H64A mutant of Phosphoketolase from Bifidobacterium Breve complexed with a tricyclic ring form of thiamine diphosphate | | Descriptor: | 1,2-ETHANEDIOL, 2-[(9aR)-2,7-dimethyl-9a,10-dihydro-5H-pyrimido[4,5-d][1,3]thiazolo[3,2-a]pyrimidin-8-yl]ethyl trihydrogen diphosphate, MAGNESIUM ION, ... | | Authors: | Suzuki, R, Katayama, T, Kim, B.-J, Wakagi, T, Shoun, H, Ashida, H, Yamamoto, K, Fushinobu, S. | | Deposit date: | 2010-04-22 | | Release date: | 2010-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structures of phosphoketolase: thiamine diphosphate-dependent dehydration mechanism
J.Biol.Chem., 285, 2010
|
|
5NAP
 
 | | Torpedo californica acetylcholinesterase in complex with a non-chiral donepezil-like inhibitor 17 | | Descriptor: | (2~{E})-5,6-dimethoxy-2-[[1-(phenylmethyl)piperidin-4-yl]methylidene]-3~{H}-inden-1-one, Acetylcholinesterase, alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Caliandro, R, Pesaresi, A, Lamba, D. | | Deposit date: | 2017-02-28 | | Release date: | 2018-05-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Kinetic and structural studies on the interactions of Torpedo californica acetylcholinesterase with two donepezil-like rigid analogues.
J Enzyme Inhib Med Chem, 33, 2018
|
|
5ECI
 
 | | Crystal Structure of FIN219-FIP1 complex with JA, ATP and Mg | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLUTATHIONE, Glutathione S-transferase U20, ... | | Authors: | Chen, C.Y, Cheng, Y.S. | | Deposit date: | 2015-10-20 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structural basis of jasmonate-amido synthetase FIN219 in complex with glutathione S-transferase FIP1 during the JA signal regulation
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6UM5
 
 | | Cryo-EM structure of HIV-1 neutralizing antibody DH270 UCA3 in complex with CH848 10.17DT Env | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Acharya, P, Henderson, R.C, Saunders, K.O, Haynes, B.F. | | Deposit date: | 2019-10-09 | | Release date: | 2019-12-18 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Targeted selection of HIV-specific antibody mutations by engineering B cell maturation.
Science, 366, 2019
|
|
3D0C
 
 | | Crystal structure of dihydrodipicolinate synthase from Oceanobacillus iheyensis at 1.9 A resolution | | Descriptor: | Dihydrodipicolinate synthase | | Authors: | Satyanarayana, L, Eswaramoorthy, S, Sauder, J.M, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-05-01 | | Release date: | 2008-05-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of dihydrodipicolinate synthase from Oceanobacillus iheyensis at 1.9 A resolution.
To be Published
|
|
