3H2H
 
 | |
2HQM
 
 | |
3H53
 
 | | Crystal Structure of human alpha-N-acetylgalactosaminidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-N-acetylgalactosaminidase, ... | | Authors: | Clark, N.E, Garman, S.C. | | Deposit date: | 2009-04-21 | | Release date: | 2009-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The 1.9 a structure of human alpha-N-acetylgalactosaminidase: The molecular basis of Schindler and Kanzaki diseases
J.Mol.Biol., 393, 2009
|
|
1LTE
 
 | | STRUCTURE OF A LEGUME LECTIN WITH AN ORDERED N-LINKED CARBOHYDRATE IN COMPLEX WITH LACTOSE | | Descriptor: | CALCIUM ION, CORAL TREE LECTIN, MANGANESE (II) ION, ... | | Authors: | Shaanan, B, Lis, H, Sharon, N. | | Deposit date: | 1991-06-25 | | Release date: | 1994-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a legume lectin with an ordered N-linked carbohydrate in complex with lactose.
Science, 254, 1991
|
|
2AHK
 
 | | Crystal structure of the met-form of the copper-bound Streptomyces castaneoglobisporus tyrosinase in complex with a caddie protein obtained by soking in cupric sulfate for 6 months | | Descriptor: | CADDIE PROTEIN ORF378, COPPER (II) ION, NITRATE ION, ... | | Authors: | Matoba, Y, Kumagai, T, Yamamoto, A, Yoshitsu, H, Sugiyama, M. | | Deposit date: | 2005-07-28 | | Release date: | 2006-01-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Crystallographic Evidence That the Dinuclear Copper Center of Tyrosinase Is Flexible during Catalysis
J.Biol.Chem., 281, 2006
|
|
3H2I
 
 | |
5IPZ
 
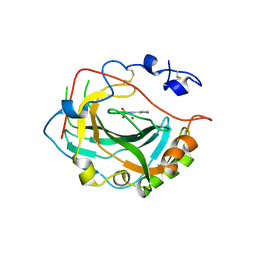 | | Crystal structure of human carbonic anhydrase isozyme IV with 5-(2-amino-1,3-thiazol-4-yl)-2-chlorobenzenesulfonamide | | Descriptor: | 5-(2-amino-1,3-thiazol-4-yl)-2-chlorobenzene-1-sulfonamide, Carbonic anhydrase 4, ZINC ION | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2016-03-10 | | Release date: | 2017-03-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Intrinsic thermodynamics of high affinity inhibitor binding to recombinant human carbonic anhydrase IV.
Eur. Biophys. J., 47, 2018
|
|
5B5K
 
 | | Crystal structure of Izumo1, the mammalian sperm ligand for egg Juno | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Izumo sperm-egg fusion protein 1 | | Authors: | Nishimura, K, Han, L, De Sanctis, D, Jovine, L. | | Deposit date: | 2016-05-11 | | Release date: | 2016-07-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structure of sperm Izumo1 reveals unexpected similarities with Plasmodium invasion proteins.
Curr.Biol., 26, 2016
|
|
4Y82
 
 | | Yeast 20S proteasome in complex with Ac-LAY-ep | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Ac-LAY-ep, CHLORIDE ION, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2015-02-16 | | Release date: | 2015-06-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Systematic Analyses of Substrate Preferences of 20S Proteasomes Using Peptidic Epoxyketone Inhibitors.
J.Am.Chem.Soc., 137, 2015
|
|
4Y8S
 
 | |
5ZLH
 
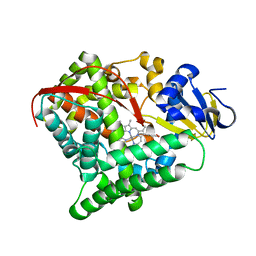 | | Crystal structure of Mn-ProtoporphyrinIX-reconstituted P450BM3 | | Descriptor: | Bifunctional cytochrome P450/NADPH--P450 reductase, MANGANESE PROTOPORPHYRIN IX | | Authors: | Omura, K, Aiba, Y, Onoda, H, Sugimoto, H, Shoji, O, Watanabe, Y. | | Deposit date: | 2018-03-28 | | Release date: | 2018-08-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Reconstitution of full-length P450BM3 with an artificial metal complex by utilising the transpeptidase Sortase A.
Chem. Commun. (Camb.), 54, 2018
|
|
4Y80
 
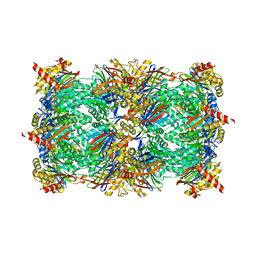 | | Yeast 20S proteasome in complex with Ac-LAI-ep | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Ac-LAI-ep, CHLORIDE ION, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2015-02-16 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Systematic Analyses of Substrate Preferences of 20S Proteasomes Using Peptidic Epoxyketone Inhibitors.
J.Am.Chem.Soc., 137, 2015
|
|
4YA7
 
 | |
5DEY
 
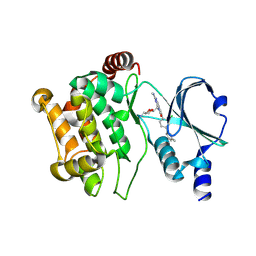 | | Crystal structure of PAK1 in complex with an inhibitor compound G-5555 | | Descriptor: | 8-[(trans-5-amino-1,3-dioxan-2-yl)methyl]-6-[2-chloro-4-(6-methylpyridin-2-yl)phenyl]-2-(methylamino)pyrido[2,3-d]pyrimidin-7(8H)-one, Serine/threonine-protein kinase PAK 1 | | Authors: | Oh, A, Tam, C, Wang, W. | | Deposit date: | 2015-08-26 | | Release date: | 2016-01-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design of Selective PAK1 Inhibitor G-5555: Improving Properties by Employing an Unorthodox Low-pK a Polar Moiety.
Acs Med.Chem.Lett., 6, 2015
|
|
7N2A
 
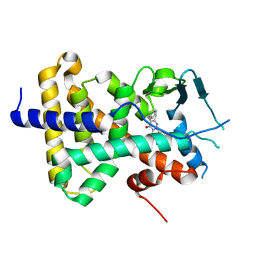 | | human PXR LBD bound to compound 2 | | Descriptor: | 5-benzyl-2-(3-fluoro-2-hydroxyphenyl)-6-methyl-3-(2-phenylethyl)pyrimidin-4(3H)-one, Isoform 1C of Nuclear receptor subfamily 1 group I member 2 | | Authors: | Williams, S.P, Wisely, G.B, Ramanjulu, J.M. | | Deposit date: | 2021-05-28 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Overcoming the Pregnane X Receptor Liability: Rational Design to Eliminate PXR-Mediated CYP Induction.
Acs Med.Chem.Lett., 12, 2021
|
|
7LRT
 
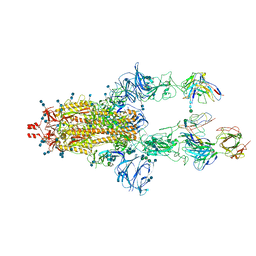 | |
4PPL
 
 | | Crystal structure of eCGP123 H193Q variant at pH 7.5 | | Descriptor: | Monomeric Azami Green | | Authors: | Don Paul, C, Traore, D.A.K, Devenish, R.J, Close, D, Bell, T, Bradbury, A, Wilce, M.C.J, Prescott, M. | | Deposit date: | 2014-02-27 | | Release date: | 2015-04-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-Ray Crystal Structure and Properties of Phanta, a Weakly Fluorescent Photochromic GFP-Like Protein.
Plos One, 10, 2015
|
|
6W69
 
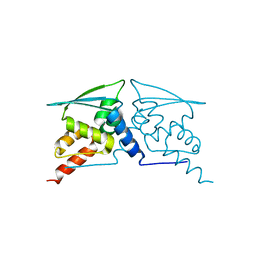 | | The structure of F64, S172A Keap1-BTB domain | | Descriptor: | Kelch-like ECH-associated protein 1 | | Authors: | Mena, E.L, Gee, C.L, Kuriyan, J, Rape, M. | | Deposit date: | 2020-03-16 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Structural basis for dimerization quality control.
Nature, 586, 2020
|
|
6T2E
 
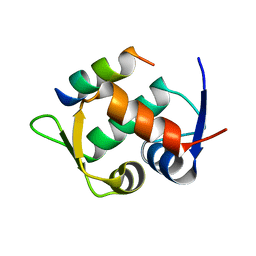 | | Multicomponent Peptide Stapling as a Diversity-Driven Tool for the Development of Inhibitors of Protein-Protein Interactions | | Descriptor: | E3 ubiquitin-protein ligase Mdm2, Stapled peptide GAR300-Gm | | Authors: | Groves, R.M, Ali, M.A, Atmaj, J, van Oosterwijk, N, Domling, A, Rivera, G.D, Ricardo, G.M. | | Deposit date: | 2019-10-08 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Multicomponent Peptide Stapling as a Diversity-Driven Tool for the Development of Inhibitors of Protein-Protein Interactions.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6T2F
 
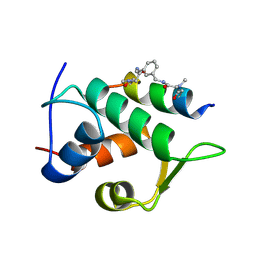 | | Multicomponent Peptide Stapling as a Diversity-Driven Tool for the Development of Inhibitors of Protein-Protein Interactions | | Descriptor: | 2-(methylamino)-~{N}-[[3-[[2-(methylamino)ethanoylamino]methyl]phenyl]methyl]ethanamide, E3 ubiquitin-protein ligase Mdm2, MDM2 in complex with GAR300-Am | | Authors: | Groves, R.M, Ali, M.A, Atmaj, J, van Oosterwijk, N, Domling, A, Rivera, G.D, Ricardo, G.M. | | Deposit date: | 2019-10-08 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Multicomponent Peptide Stapling as a Diversity-Driven Tool for the Development of Inhibitors of Protein-Protein Interactions.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6W68
 
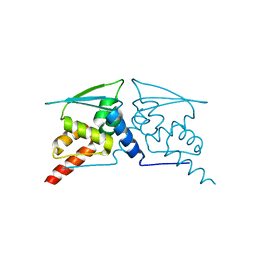 | |
7KVN
 
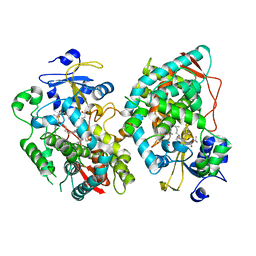 | | Human CYP3A4 bound to an inhibitor | | Descriptor: | Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE, tert-butyl [(2S)-1-(1H-indol-3-yl)-3-{[(2R)-1-oxo-3-phenyl-1-{[3-(pyridin-3-yl)propyl]amino}propan-2-yl]sulfanyl}propan-2-yl]carbamate | | Authors: | Sevrioukova, I. | | Deposit date: | 2020-11-28 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Rational Design of CYP3A4 Inhibitors: A One-Atom Linker Elongation in Ritonavir-Like Compounds Leads to a Marked Improvement in the Binding Strength.
Int J Mol Sci, 22, 2021
|
|
7KVK
 
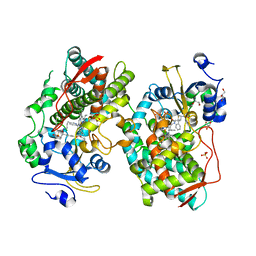 | | Human CYP3A4 bound to an inhibitor | | Descriptor: | 1,2-ETHANEDIOL, Cytochrome P450 3A4, GLYCEROL, ... | | Authors: | Sevrioukova, I. | | Deposit date: | 2020-11-28 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Rational Design of CYP3A4 Inhibitors: A One-Atom Linker Elongation in Ritonavir-Like Compounds Leads to a Marked Improvement in the Binding Strength.
Int J Mol Sci, 22, 2021
|
|
3H2G
 
 | |
7KVH
 
 | | Human CYP3A4 bound to an inhibitor | | Descriptor: | Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE, tert-butyl [(2S)-1-{[(2R)-1-oxo-3-phenyl-1-{[3-(pyridin-3-yl)propyl]amino}propan-2-yl]sulfanyl}-3-phenylpropan-2-yl]carbamate | | Authors: | Sevrioukova, I. | | Deposit date: | 2020-11-28 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.791 Å) | | Cite: | Rational Design of CYP3A4 Inhibitors: A One-Atom Linker Elongation in Ritonavir-Like Compounds Leads to a Marked Improvement in the Binding Strength.
Int J Mol Sci, 22, 2021
|
|
