6TUK
 
 | | Crystal structure of Fdr9 | | Descriptor: | (R,R)-2,3-BUTANEDIOL, FLAVIN-ADENINE DINUCLEOTIDE, Putative oxidoreductase, ... | | Authors: | Rodriguez, A, Kluenemann, T, Blankenfeldt, W, Schallmey, A. | | Deposit date: | 2020-01-07 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Expression, purification and crystal structure determination of a ferredoxin reductase from the actinobacterium Thermobifida fusca.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
5MLT
 
 | |
5MM9
 
 | | VIM-2_2b. Metallo-beta-Lactamase Inhibitors by Bioisosteric Replacement: Preparation, Activity and Binding | | Descriptor: | (2~{R})-2-diethoxyphosphoryl-5-phenyl-pentane-1-thiol, MAGNESIUM ION, Metallo-beta-lactamase VIM-17, ... | | Authors: | Skagseth, S, Akhter, S, Paulsen, M.H, Samuelsen, O, Muhammad, Z, Leiros, H.-K.S, Bayer, A. | | Deposit date: | 2016-12-08 | | Release date: | 2017-03-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Metallo-beta-lactamase inhibitors by bioisosteric replacement: Preparation, activity and binding.
Eur J Med Chem, 135, 2017
|
|
6T60
 
 | |
6T6C
 
 | | Complex with chitin oligomer of C-type lysozyme from the upper gastrointestinal tract of Opisthocomus hoatzin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Lysozyme C | | Authors: | Taylor, E.J, Skjot, M, Skov, L.K, Klausen, M, De Maria, L, Gippert, G.P, Turkenburg, J.P, Davies, G.J, Wilson, K.S. | | Deposit date: | 2019-10-18 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | The C-Type Lysozyme from the upper Gastrointestinal Tract of Opisthocomus hoatzin, the Stinkbird.
Int J Mol Sci, 20, 2019
|
|
6TSY
 
 | | Cytochrome c6 from Thermosynechococcus elongatus in a monoclinic space group | | Descriptor: | Cytochrome c6, HEME C, SULFATE ION | | Authors: | Feiler, C.G, Falke, S, Sarrou, I. | | Deposit date: | 2019-12-21 | | Release date: | 2020-01-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structures of native cytochrome c6from Thermosynechococcus elongatus in two different space groups and implications for its oligomerization.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
6T6N
 
 | | Crystal structure of Klebsiella pneumoniae FabG2(NADH-dependent) in complex with NADH at 2.5 A resolution | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 3-oxoacyl-[acyl-carrier protein] reductase, D-MALATE, ... | | Authors: | Vella, P, Schnell, R, Lindqvist, Y, Schneider, G. | | Deposit date: | 2019-10-18 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A FabG inhibitor targeting an allosteric binding site inhibits several orthologs from Gram-negative ESKAPE pathogens.
Bioorg.Med.Chem., 30, 2021
|
|
5OOA
 
 | | Streptomyces PAC13 (Y89F) with uridine | | Descriptor: | Putative cupin_2 domain-containing isomerase, URIDINE | | Authors: | Chung, C, Michailidou, F. | | Deposit date: | 2017-08-06 | | Release date: | 2017-08-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Pac13 is a Small, Monomeric Dehydratase that Mediates the Formation of the 3'-Deoxy Nucleoside of Pacidamycins.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5OOY
 
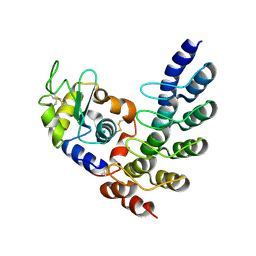 | | Designed Ankyrin Repeat Protein (DARPin) VHAH-1 in complex with Lysozyme | | Descriptor: | DARPin VHAH-1, Lysozyme C | | Authors: | Hogan, B.J, Fischer, G, Houlihan, G, Edmond, S, Huovinen, T.T.K, Hollfelder, F, Hyvonen, M. | | Deposit date: | 2017-08-09 | | Release date: | 2018-08-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Designed Ankyrin Repeat Protein (DARPin) VHAH-1 in complex with Lysozyme
To be published
|
|
6TTR
 
 | | Crystal Structure of the coiled coil and GGDEF domain of DgcB from Caulobacter crescentus in complex with c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), GGDEF diguanylate cyclase DgcB, PHOSPHATE ION | | Authors: | Holzschuh, F, Schirmer, T, Teixeira, R.D. | | Deposit date: | 2019-12-30 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structure of the coiled coil and GGDEF domain of DgcB from Caulobacter crescentus in complex with c-di-GMP
To Be Published
|
|
6TCK
 
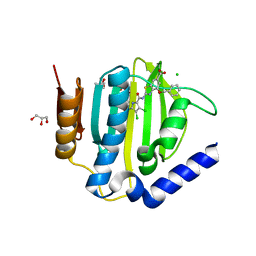 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with ULD-2 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-[[3,4-bis(chloranyl)-5-methyl-1~{H}-pyrrol-2-yl]carbonylamino]-4-phenylmethoxy-1,3-benzothiazole-6-carboxylic acid, CALCIUM ION, ... | | Authors: | Welin, M, Kimbung, R, Focht, D. | | Deposit date: | 2019-11-06 | | Release date: | 2020-09-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Rational design of balanced dual-targeting antibiotics with limited resistance.
Plos Biol., 18, 2020
|
|
6TCQ
 
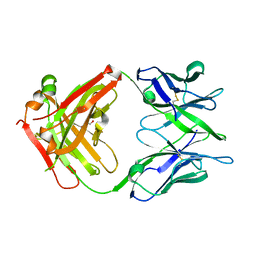 | | Crystal structure of the omalizumab Fab Ser81Arg and Gln83Arg light chain mutant | | Descriptor: | GLYCEROL, Omalizumab Fab Ser81Arg and Gln83Arg light chain mutant | | Authors: | Mitropoulou, A.N, Ceska, T, Beavil, A.J, Henry, A.J, McDonnell, J.M, Sutton, B.J, Davies, A.M. | | Deposit date: | 2019-11-06 | | Release date: | 2020-03-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Engineering the Fab fragment of the anti-IgE omalizumab to prevent Fab crystallization and permit IgE-Fc complex crystallization.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
6TDO
 
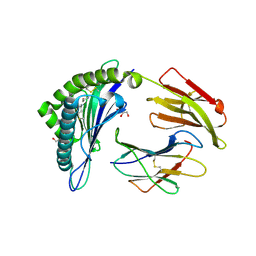 | | Crystal structure of the disulfide engineered HLA-A0201 molecule in complex with one GM dipeptide in the A pocket. | | Descriptor: | Beta-2-microglobulin, GLYCEROL, GLYCINE, ... | | Authors: | Anjanappa, R, Garcia Alai, M, Springer, S, Meijers, R. | | Deposit date: | 2019-11-09 | | Release date: | 2020-03-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structures of peptide-free and partially loaded MHC class I molecules reveal mechanisms of peptide selection.
Nat Commun, 11, 2020
|
|
6TDQ
 
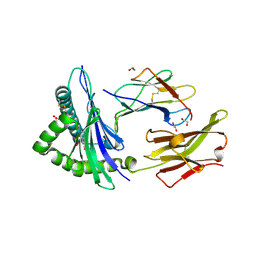 | | Crystal structure of the disulfide engineered HLA-A0201 molecule in complex with one GM dipeptide in the A pocket and one GM dipeptide in the F pocket. | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, CHLORIDE ION, ... | | Authors: | Anjanappa, R, Garcia Alai, M, Springer, S, Meijers, R. | | Deposit date: | 2019-11-10 | | Release date: | 2020-03-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of peptide-free and partially loaded MHC class I molecules reveal mechanisms of peptide selection.
Nat Commun, 11, 2020
|
|
5ONJ
 
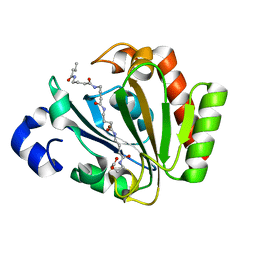 | | YnDL in Complex with 5 amino acid (PGA) complex | | Descriptor: | (2R,3S,4R,5R,6R)-6-((1R,2R,3S,4R,6S)-4,6-DIAMINO-2,3-DIHYDROXYCYCLOHEXYLOXY)-5-AMINO-2-(AMINOMETHYL)-TETRAHYDRO-2H-PYRAN-3,4-DIOL, YndL, ZINC ION | | Authors: | Ramaswamy, S, Rasheed, M, Morelli, C, Calvio, C, Sutton, B, Pastore, A. | | Deposit date: | 2017-08-03 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structure of PghL hydrolase bound to its substrate poly-gamma-glutamate.
Febs J., 285, 2018
|
|
6TDS
 
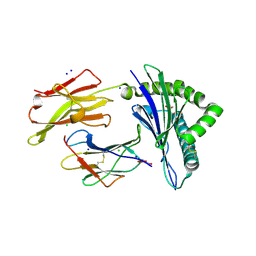 | | Crystal structure of the disulfide engineered HLA-A0201 molecule without peptide bound after NaCl wash | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, CHLORIDE ION, ... | | Authors: | Anjanappa, R, Garcia Alai, M, Springer, S, Meijers, R. | | Deposit date: | 2019-11-10 | | Release date: | 2020-03-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of peptide-free and partially loaded MHC class I molecules reveal mechanisms of peptide selection.
Nat Commun, 11, 2020
|
|
5ONP
 
 | |
5OQ3
 
 | | High resolution structure of the functional region of Cwp19 from Clostridium difficile | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Cwp19, ... | | Authors: | Bradshaw, W.J, Kirby, J.M, Roberts, A.K, Shone, C.C, Acharya, K.R. | | Deposit date: | 2017-08-10 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The molecular structure of the glycoside hydrolase domain of Cwp19 from Clostridium difficile.
FEBS J., 284, 2017
|
|
6TG6
 
 | | Toprim domain of RNase M5 | | Descriptor: | Ribonuclease M5 | | Authors: | Oerum, S, Catala, M, Tisne, C. | | Deposit date: | 2019-11-15 | | Release date: | 2020-09-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structures of B. subtilis Maturation RNases Captured on 50S Ribosome with Pre-rRNAs.
Mol.Cell, 80, 2020
|
|
5ON7
 
 | | The ENTH domain from epsin-2 in complex with phosphatidylinositol 4,5-bisphosphate (PIP2) | | Descriptor: | Epsin-2, SULFATE ION, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate | | Authors: | Garcia-Alai, M, GIeras, A, Meijers, R. | | Deposit date: | 2017-08-03 | | Release date: | 2018-03-07 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Epsin and Sla2 form assemblies through phospholipid interfaces.
Nat Commun, 9, 2018
|
|
5OWL
 
 | | Low salt structure of human protein kinase CK2alpha in complex with 3-aminopropyl-4,5,6,7-tetrabromobenzimidazol | | Descriptor: | 3-[4,5,6,7-tetrakis(bromanyl)benzimidazol-1-yl]propan-1-amine, Casein kinase II subunit alpha, GLYCEROL, ... | | Authors: | Niefind, K, Bretner, M, Chojnacki, C. | | Deposit date: | 2017-09-01 | | Release date: | 2018-07-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Biological properties and structural study of new aminoalkyl derivatives of benzimidazole and benzotriazole, dual inhibitors of CK2 and PIM1 kinases.
Bioorg. Chem., 80, 2018
|
|
6TC7
 
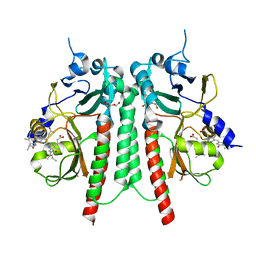 | | PAS-GAF bidomain of Glycine max phytochromeA | | Descriptor: | DI(HYDROXYETHYL)ETHER, PHYCOCYANOBILIN, Phytochrome | | Authors: | Nagano, S, Guan, K, Shenkutie, S.M, Hughes, J.E. | | Deposit date: | 2019-11-05 | | Release date: | 2020-05-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structural insights into photoactivation and signalling in plant phytochromes.
Nat.Plants, 6, 2020
|
|
5OO4
 
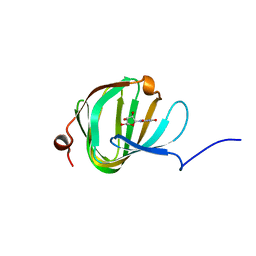 | | Streptomyces PAC13 with uridine | | Descriptor: | Putative cupin_2 domain-containing isomerase, URIDINE | | Authors: | Chung, C, Michailidou, F. | | Deposit date: | 2017-08-05 | | Release date: | 2017-08-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Pac13 is a Small, Monomeric Dehydratase that Mediates the Formation of the 3'-Deoxy Nucleoside of Pacidamycins.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
6TCI
 
 | |
5OXF
 
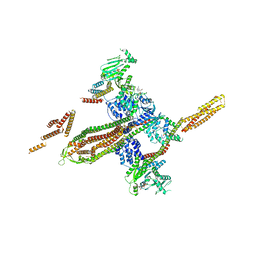 | |
