3FMH
 
 | |
8RR6
 
 | |
8RSM
 
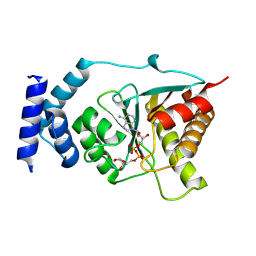 | | Crystal structure of Streptococcus pyogenes macrodomain in complex with ADP-ribose | | Descriptor: | Protein-ADP-ribose hydrolase, ZINC ION, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Ariza, A. | | Deposit date: | 2024-01-24 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Evolutionary and molecular basis of ADP-ribosylation reversal by zinc-dependent macrodomains.
J.Biol.Chem., 300, 2024
|
|
4E6A
 
 | | p38a-PIA23 complex | | Descriptor: | (2S)-2-methoxy-3-(octadecyloxy)propyl (1R,2R,3R,4S,6S)-2,3,4-trihydroxy-6-(2-methylpropoxy)cyclohexyl hydrogen (S)-phosphate, Mitogen-activated protein kinase 14 | | Authors: | Livnah, O, Tzarum, N, Eisenberg-Domovich, Y. | | Deposit date: | 2012-03-15 | | Release date: | 2012-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Lipid Molecules Induce p38 alpha Activation via a Novel Molecular Switch.
J.Mol.Biol., 424, 2012
|
|
4UNP
 
 | | Mtb TMK in complex with compound 34 | | Descriptor: | 5-methyl-7-propyl-1,6-naphthyridin-2(1H)-one, THYMIDYLATE KINASE | | Authors: | Read, J.A, Hussein, S, Gingell, H, Tucker, J. | | Deposit date: | 2014-05-30 | | Release date: | 2015-01-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure Guided Lead Generation for M. Tuberculosis Thymidylate Kinase (Mtb Tmk): Discovery of 3-Cyanopyridone and 1,6-Naphthyridin-2-One as Potent Inhibitors.
J.Med.Chem., 58, 2015
|
|
4UMW
 
 | | CRYSTAL STRUCTURE OF A ZINC-TRANSPORTING PIB-TYPE ATPASE IN E2.PI STATE | | Descriptor: | MAGNESIUM ION, TETRAFLUOROALUMINATE ION, ZINC-TRANSPORTING ATPASE | | Authors: | Wang, K.T, Sitsel, O, Meloni, G, Autzen, H.E, Andersson, M, Klymchuk, T, Nielsen, A.M, Rees, D.C, Nissen, P, Gourdon, P. | | Deposit date: | 2014-05-21 | | Release date: | 2014-08-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.705 Å) | | Cite: | Structure and Mechanism of Zn(2+)-Transporting P-Type Atpases.
Nature, 514, 2014
|
|
3NOV
 
 | | Crystal Structure of D17E Isocyanide Hydratase from Pseudomonas fluorescens | | Descriptor: | ACETATE ION, ThiJ/PfpI family protein | | Authors: | Lakshminarasimhan, M, Madzelan, P, Nan, R, Milkovic, N.M, Wilson, M.A. | | Deposit date: | 2010-06-25 | | Release date: | 2010-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Evolution of New Enzymatic Function by Structural Modulation of Cysteine Reactivity in Pseudomonas fluorescens Isocyanide Hydratase.
J.Biol.Chem., 285, 2010
|
|
8RSK
 
 | | Crystal structure of Methanobrevibacter oralis macrodomain in complex with Asn-ADPr | | Descriptor: | (2~{S})-4-[[(2~{S},3~{R},4~{S},5~{R})-5-[[[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxymethyl]-3,4-bis(oxidanyl)oxolan-2-yl]amino]-2-azanyl-4-oxidanylidene-butanoic acid, ACETATE ION, GLYCEROL, ... | | Authors: | Ariza, A. | | Deposit date: | 2024-01-24 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.359 Å) | | Cite: | Evolutionary and molecular basis of ADP-ribosylation reversal by zinc-dependent macrodomains.
J.Biol.Chem., 300, 2024
|
|
8R9T
 
 | |
4UNQ
 
 | | Mtb TMK in complex with compound 36 | | Descriptor: | 4-[(R)-methylsulfinyl]-2-oxo-6-[3-(trifluoromethoxy)phenyl]-1,2-dihydropyridine-3-carbonitrile, SODIUM ION, THYMIDYLATE KINASE | | Authors: | Read, J.A, Hussein, S, Gingell, H, Tucker, J. | | Deposit date: | 2014-05-30 | | Release date: | 2015-06-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure Guided Lead Generation for M. Tuberculosis Thymidylate Kinase (Mtb Tmk): Discovery of 3-Cyanopyridone and 1,6-Naphthyridin-2-One as Potent Inhibitors.
J.Med.Chem., 58, 2015
|
|
4UNS
 
 | | Mtb TMK in complex with compound 40 | | Descriptor: | N-[4-(3-CYANO-7-ETHYL-5-METHYL-2-OXO-1H-1,6-NAPHTHYRIDIN-4-YL)PHENYL]METHANESULFONAMIDE, SODIUM ION, THYMIDYLATE KINASE | | Authors: | Read, J.A, Hussein, S, Gingell, H, Tucker, J. | | Deposit date: | 2014-05-30 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structure Guided Lead Generation for M. Tuberculosis Thymidylate Kinase (Mtb Tmk): Discovery of 3-Cyanopyridone and 1,6-Naphthyridin-2-One as Potent Inhibitors.
J.Med.Chem., 58, 2015
|
|
1ZUG
 
 | | STRUCTURE OF PHAGE 434 CRO PROTEIN, NMR, 20 STRUCTURES | | Descriptor: | PHAGE 434 CRO PROTEIN | | Authors: | Padmanabhan, S, Jimenez, M.A, Gonzalez, C, Sanz, J.M, Gimenez-Gallego, G, Rico, M. | | Deposit date: | 1997-03-14 | | Release date: | 1997-07-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure and stability of phage 434 Cro protein.
Biochemistry, 36, 1997
|
|
8RSJ
 
 | |
4UOH
 
 | | Crystallographic structure of nucleoside diphosphate kinase from Litopenaeus vannamei complexed with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, NUCLEOSIDE DIPHOSPHATE KINASE | | Authors: | Lopez-Zavala, A.A, Guevara-Hernandez, E, Stojanoff, V, Rudino-Pinera, E, Sotelo-Mundo, R.R. | | Deposit date: | 2014-06-03 | | Release date: | 2014-09-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.007 Å) | | Cite: | Structure of Nucleoside Diphosphate Kinase from Pacific Shrimp (Litopenaeus Vannamei) in Binary Complexes with Purine and Pyrimidine Nucleoside Diphosphates
Acta Crystallogr.,Sect.F, 79, 2014
|
|
8RSL
 
 | | Crystal structure of Staphylococcus aureus macrodomain | | Descriptor: | Protein-ADP-ribose hydrolase, ZINC ION | | Authors: | Ariza, A. | | Deposit date: | 2024-01-24 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Evolutionary and molecular basis of ADP-ribosylation reversal by zinc-dependent macrodomains.
J.Biol.Chem., 300, 2024
|
|
8RSI
 
 | | Crystal structure of Methanobrevibacter oralis macrodomain | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTAMIC ACID, ... | | Authors: | Ariza, A. | | Deposit date: | 2024-01-24 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.059 Å) | | Cite: | Evolutionary and molecular basis of ADP-ribosylation reversal by zinc-dependent macrodomains.
J.Biol.Chem., 300, 2024
|
|
8RSN
 
 | |
3IFJ
 
 | |
8AO6
 
 | | electrophilic inhibitor (7) of ERK2 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Mitogen-activated protein kinase 1, ... | | Authors: | Cleasby, A. | | Deposit date: | 2022-08-08 | | Release date: | 2022-09-28 | | Last modified: | 2022-10-05 | | Method: | X-RAY DIFFRACTION (1.811 Å) | | Cite: | X-ray Screening of an Electrophilic Fragment Library and Application toward the Development of a Novel ERK 1/2 Covalent Inhibitor.
J.Med.Chem., 65, 2022
|
|
8AO8
 
 | | Specific covalent inhibitor(9) of ERK2 | | Descriptor: | 1,2-ETHANEDIOL, 1-[(2~{R})-2-(3-methylimidazol-4-yl)piperidin-1-yl]propan-1-one, Mitogen-activated protein kinase 1, ... | | Authors: | Cleasby, A. | | Deposit date: | 2022-08-08 | | Release date: | 2022-09-28 | | Last modified: | 2023-05-24 | | Method: | X-RAY DIFFRACTION (1.697 Å) | | Cite: | X-ray Screening of an Electrophilic Fragment Library and Application toward the Development of a Novel ERK 1/2 Covalent Inhibitor.
J.Med.Chem., 65, 2022
|
|
3FLN
 
 | | P38 kinase crystal structure in complex with R1487 | | Descriptor: | 6-(2,4-difluorophenoxy)-8-methyl-2-(tetrahydro-2H-pyran-4-ylamino)pyrido[2,3-d]pyrimidin-7(8H)-one, Mitogen-activated protein kinase 14 | | Authors: | Kuglstatter, A, Knapp, M. | | Deposit date: | 2008-12-19 | | Release date: | 2009-12-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Discovery of Pamapimod, R1503 and R1487 as Orally Bioavailable and Highly Selective Inhibitors of p38 Map Kinase
To be Published
|
|
8AO5
 
 | | Specific covalent inhibitor (6) of ERK2 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Mitogen-activated protein kinase 1, SULFATE ION, ... | | Authors: | Cleasby, A. | | Deposit date: | 2022-08-08 | | Release date: | 2022-09-28 | | Last modified: | 2022-10-05 | | Method: | X-RAY DIFFRACTION (1.595 Å) | | Cite: | X-ray Screening of an Electrophilic Fragment Library and Application toward the Development of a Novel ERK 1/2 Covalent Inhibitor.
J.Med.Chem., 65, 2022
|
|
8AOB
 
 | | Specific covalent inhibitor(12) of ERK2 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Mitogen-activated protein kinase 1, SULFATE ION, ... | | Authors: | Cleasby, A. | | Deposit date: | 2022-08-08 | | Release date: | 2022-09-28 | | Last modified: | 2022-10-05 | | Method: | X-RAY DIFFRACTION (1.623 Å) | | Cite: | X-ray Screening of an Electrophilic Fragment Library and Application toward the Development of a Novel ERK 1/2 Covalent Inhibitor.
J.Med.Chem., 65, 2022
|
|
8AOF
 
 | | Specific covalent inhibitor(16) of ERK2 | | Descriptor: | 1,2-ETHANEDIOL, Mitogen-activated protein kinase 1, SULFATE ION, ... | | Authors: | Cleasby, A. | | Deposit date: | 2022-08-08 | | Release date: | 2022-09-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.615 Å) | | Cite: | X-ray Screening of an Electrophilic Fragment Library and Application toward the Development of a Novel ERK 1/2 Covalent Inhibitor.
J.Med.Chem., 65, 2022
|
|
3FLY
 
 | |
