6A28
 
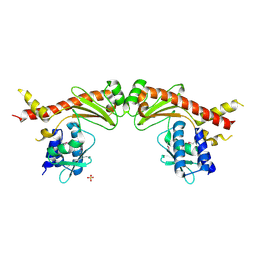 | | Crystal structure of PprA W183R mutant form 2 | | Descriptor: | DNA repair protein PprA, SULFATE ION | | Authors: | Adachi, M, Shibazaki, C, Shimizu, R, Arai, S, Satoh, K, Narumi, I, Kuroki, R. | | Deposit date: | 2018-06-09 | | Release date: | 2018-12-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.193 Å) | | Cite: | Extended structure of pleiotropic DNA repair-promoting protein PprA from Deinococcus radiodurans.
FASEB J., 33, 2019
|
|
3H9E
 
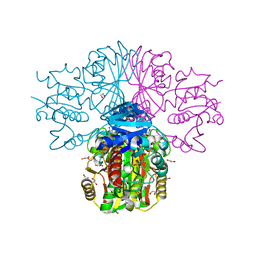 | | Crystal structure of human sperm-specific glyceraldehyde-3-phosphate dehydrogenase (GAPDS) complex with NAD and phosphate | | Descriptor: | 1,2-ETHANEDIOL, Glyceraldehyde-3-phosphate dehydrogenase, testis-specific, ... | | Authors: | Chaikuad, A, Shafqat, N, Yue, W, Cocking, R, Bray, J.E, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-04-30 | | Release date: | 2009-05-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structure and kinetic characterization of human sperm-specific glyceraldehyde-3-phosphate dehydrogenase, GAPDS.
Biochem.J., 435, 2011
|
|
5ZU3
 
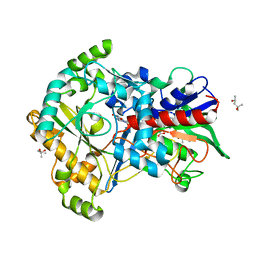 | | Effect of mutation (R554K) on FAD modification in Aspergillus oryzae RIB40formate oxidase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, Formate oxidase, ... | | Authors: | Mikami, B, Uchida, H, Doubayashi, D. | | Deposit date: | 2018-05-06 | | Release date: | 2019-05-22 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The microenvironment surrounding FAD mediates its conversion to 8-formyl-FAD in Aspergillus oryzae RIB40 formate oxidase.
J.Biochem., 166, 2019
|
|
6A2J
 
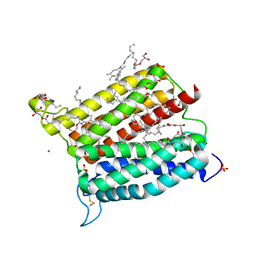 | | Crystal structure of heme A synthase from Bacillus subtilis | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, COPPER (II) ION, Heme A synthase, ... | | Authors: | Niwa, S, Takeda, K, Kosugi, M, Tsutsumi, E, Miki, K. | | Deposit date: | 2018-06-12 | | Release date: | 2018-11-21 | | Last modified: | 2018-12-12 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of heme A synthase fromBacillus subtilis.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4P7L
 
 | | Structure of Escherichia coli PgaB C-terminal domain, P212121 crystal form | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Poly-beta-1,6-N-acetyl-D-glucosamine N-deacetylase | | Authors: | Little, D.J, Li, G, Ing, C, DiFrancesco, B, Bamford, N.C, Robinson, H, Nitz, M, Pomes, R, Howell, P.L. | | Deposit date: | 2014-03-27 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Modification and periplasmic translocation of the biofilm exopolysaccharide poly-beta-1,6-N-acetyl-D-glucosamine.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2ILU
 
 | |
6I4T
 
 | | Crystal structure of the disease-causing I445M mutant of the human dihydrolipoamide dehydrogenase | | Descriptor: | Dihydrolipoyl dehydrogenase, mitochondrial, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Szabo, E, Wilk, P, Zambo, Z, Torocsik, B, Weiss, M.S, Adam-Vizi, V, Ambrus, A. | | Deposit date: | 2018-11-10 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.823 Å) | | Cite: | Underlying molecular alterations in human dihydrolipoamide dehydrogenase deficiency revealed by structural analyses of disease-causing enzyme variants.
Hum.Mol.Genet., 28, 2019
|
|
2IPG
 
 | | Crystal structure of 17alpha-hydroxysteroid dehydrogenase mutant K31A in complex with NADP+ and epi-testosterone | | Descriptor: | (10ALPHA,13ALPHA,14BETA,17ALPHA)-17-HYDROXYANDROST-4-EN-3-ONE, 1,2-ETHANEDIOL, 3(17)alpha-hydroxysteroid dehydrogenase, ... | | Authors: | Faucher, F, Cantin, L, Pereira de Jesus-Tran, K, Lemieux, M, Luu-the, V, Labrie, F, Breton, R. | | Deposit date: | 2006-10-12 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mouse 17alpha-Hydroxysteroid Dehydrogenase (AKR1C21) Binds Steroids Differently from other Aldo-keto Reductases: Identification and Characterization of Amino Acid Residues Critical for Substrate Binding.
J.Mol.Biol., 369, 2007
|
|
3EJQ
 
 | | Golgi alpha-Mannosidase II in complex with 5-substitued swainsonine analog: (5R)-5-[2'-oxo-2'-(4-methylphenyl)ethyl]-swainsonine | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 1-(4-methylphenyl)-2-[(1S,2R,5R,8R,8aR)-1,2,8-trihydroxyoctahydroindolizin-5-yl]ethanone, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kuntz, D.A, Rose, D.R. | | Deposit date: | 2008-09-18 | | Release date: | 2009-10-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural Investigation of the Binding of 5-Substituted Swainsonine Analogues to Golgi alpha-Mannosidase II.
Chembiochem, 11, 2010
|
|
5XGW
 
 | |
6A4G
 
 | | Mandelate oxidase mutant-Y128F with the monooxide FMN adduct | | Descriptor: | 1-deoxy-1-[(4aS)-4a-hydroxy-7,8-dimethyl-2,4-dioxo-3,4,4a,5-tetrahydrobenzo[g]pteridin-10(2H)-yl]-5-O-phosphono-D-ribitol, 4-hydroxymandelate oxidase | | Authors: | Li, T.L, Lin, K.H. | | Deposit date: | 2018-06-19 | | Release date: | 2019-06-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural and chemical trapping of flavin-oxide intermediates reveals substrate-directed reaction multiplicity.
Protein Sci., 29, 2020
|
|
5XTJ
 
 | | Mannanase(RmMan134A) | | Descriptor: | Endo beta-1,4-mannanase | | Authors: | Jiang, Z.Q, You, X, Huang, P. | | Deposit date: | 2017-06-19 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structural and biochemical insights into the substrate-binding mechanism of a novel glycoside hydrolase family 134 beta-mannanase.
Biochim Biophys Acta Gen Subj, 1862, 2018
|
|
6A4N
 
 | | HEWL crystals soaked in 2.5M GuHCl for 8 minutes | | Descriptor: | CHLORIDE ION, GLYCEROL, GUANIDINE, ... | | Authors: | Tushar, R, Kini, R.M, Koh, C.Y, Hosur, M.V. | | Deposit date: | 2018-06-20 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | X-ray crystallographic analysis of time-dependent binding of guanidine hydrochloride to HEWL: First steps during protein unfolding.
Int. J. Biol. Macromol., 122, 2019
|
|
2IUK
 
 | | Crystal structure of Soybean Lipoxygenase-D | | Descriptor: | FE (III) ION, SEED LIPOXYGENASE | | Authors: | Youn, B, Sellhorn, G.E, Mirchel, R.J, Gaffney, B.J, Grimes, H.D, Kang, C. | | Deposit date: | 2006-06-05 | | Release date: | 2006-10-11 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of Vegetative Soybean Lipoxygenase Vlx-B and Vlx-D, and Comparisons with Seed Isoforms Lox-1 and Lox-3.
Proteins, 65, 2006
|
|
3EF9
 
 | | Replacement of Val3 in Human Thymidylate Synthase Affects Its Kinetic Properties and Intracellular Stability | | Descriptor: | SULFATE ION, Thymidylate synthase | | Authors: | Huang, X, Gibson, L.M, Bell, B.J, Lovelace, L.L, Pena, M.M, Berger, F.G, Berger, S.H. | | Deposit date: | 2008-09-08 | | Release date: | 2010-03-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Replacement of Val3 in human thymidylate synthase affects its kinetic properties and intracellular stability .
Biochemistry, 49, 2010
|
|
7Q3X
 
 | |
6IK6
 
 | | Crystal structure of Tomato beta-galactosidase (TBG) 4 with beta-1,4-galactobiose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-galactosidase, ... | | Authors: | Matsuyama, K, Nakae, S, Igarashi, K, Tada, T, Ishimaru, M. | | Deposit date: | 2018-10-15 | | Release date: | 2018-11-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.791 Å) | | Cite: | Substrate-recognition mechanism of tomato beta-galactosidase 4 using X-ray crystallography and docking simulation.
Planta, 252, 2020
|
|
4KUG
 
 | |
4PHU
 
 | | Crystal structure of Human GPR40 bound to allosteric agonist TAK-875 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, DIMETHYL SULFOXIDE, Free fatty acid receptor 1,Lysozyme, ... | | Authors: | Srivastava, A, Yano, J.K, Hirozane, Y, Kefala, G, Snell, G, Lane, W, Gruswitz, F, Ivetac, A, Aertgeerts, K, Nguyen, J, Jennings, A, Okada, K. | | Deposit date: | 2014-05-07 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.332 Å) | | Cite: | High-resolution structure of the human GPR40 receptor bound to allosteric agonist TAK-875.
Nature, 513, 2014
|
|
3HAZ
 
 | | Crystal structure of bifunctional proline utilization A (PutA) protein | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Tanner, J.J. | | Deposit date: | 2009-05-03 | | Release date: | 2010-02-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the bifunctional proline utilization A flavoenzyme from Bradyrhizobium japonicum
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4KVK
 
 | | Crystal structure of Oryza sativa fatty acid alpha-dioxygenase | | Descriptor: | CALCIUM ION, CHLORIDE ION, DECYL-BETA-D-MALTOPYRANOSIDE, ... | | Authors: | Zhu, G, Koszelak-Rosenblum, M, Malkowski, M.G. | | Deposit date: | 2013-05-22 | | Release date: | 2013-08-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal structures of alpha-dioxygenase from Oryza sativa: Insights into substrate binding and activation by hydrogen peroxide.
Protein Sci., 22, 2013
|
|
4PJ2
 
 | | Crystal structure of Aeromonas hydrophila PliI in complex with Meretrix lusoria lysozyme | | Descriptor: | GLYCEROL, Lysozyme, MAGNESIUM ION, ... | | Authors: | Leysen, S, Van Herreweghe, J.M, Yoneda, K, Ogata, M, Usui, T, Michiels, C.W, Araki, T, Strelkov, S.V. | | Deposit date: | 2014-05-10 | | Release date: | 2015-02-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | The structure of the proteinaceous inhibitor PliI from Aeromonas hydrophila in complex with its target lysozyme.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
2RAB
 
 | | Structure of glutathione amide reductase from Chromatium gracile in complex with NAD | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, NICKEL (II) ION, ... | | Authors: | Van Petegem, F, De Vos, D, Savvides, S, Vergauwen, B, Van Beeumen, J. | | Deposit date: | 2007-09-14 | | Release date: | 2008-02-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Understanding nicotinamide dinucleotide cofactor and substrate specificity in class I flavoprotein disulfide oxidoreductases: crystallographic analysis of a glutathione amide reductase.
J.Mol.Biol., 374, 2007
|
|
5XJN
 
 | | cytochrome P450 CREJ in complex with (4-ethylphenyl) dihydrogen phosphate | | Descriptor: | (4-ethylphenyl) dihydrogen phosphate, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Dong, S, Du, L, Li, S, Feng, Y. | | Deposit date: | 2017-05-03 | | Release date: | 2017-07-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Selective oxidation of aliphatic C-H bonds in alkylphenols by a chemomimetic biocatalytic system
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
2RAY
 
 | |
