7O3R
 
 | |
7NLA
 
 | |
7O2S
 
 | | Crystal structure of a tetrameric form of Carbonic anhydrase from Schistosoma mansoni | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Carbonic anhydrase, ... | | Authors: | Ferraroni, M, Angeli, A. | | Deposit date: | 2021-03-31 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural Insights into Schistosoma mansoni Carbonic Anhydrase (SmCA) Inhibition by Selenoureido-Substituted Benzenesulfonamides.
J.Med.Chem., 64, 2021
|
|
4MNY
 
 | | Crystal structure of urokinase-type plasminogen activator (uPA) complexed with bicyclic peptide UK903 | | Descriptor: | ACETATE ION, GLYCEROL, N,N',N''-benzene-1,3,5-triyltris(2-bromoacetamide), ... | | Authors: | Chen, S, Pojer, F, Heinis, C. | | Deposit date: | 2013-09-11 | | Release date: | 2014-02-05 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Peptide ligands stabilized by small molecules.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
8SGI
 
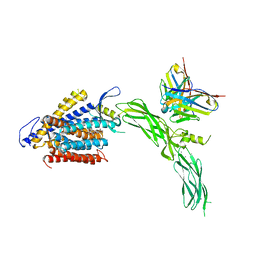 | | Cryo-EM structure of human NCX1 in complex with SEA0400 | | Descriptor: | 2-{4-[(2,5-difluorophenyl)methoxy]phenoxy}-5-ethoxyaniline, CALCIUM ION, Fab heavy chain, ... | | Authors: | Xue, J, Jiang, Y. | | Deposit date: | 2023-04-12 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural mechanisms of human cardiac sodium calcium exchanger NCX1
To Be Published
|
|
6JWU
 
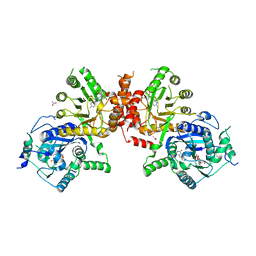 | | Crystal structure of Plasmodium falciparum HPPK-DHPS wild type with STZ-DHP | | Descriptor: | 2-AMINO-6-HYDROXYMETHYL-7,8-DIHYDRO-3H-PTERIDIN-4-ONE, 4-{[(2-amino-4-oxo-3,4,7,8-tetrahydropteridin-6-yl)methyl]amino}-N-(1,3-thiazol-2-yl)benzenesulfonamide, 7,8-dihydro-6-hydroxymethylpterin pyrophosphokinase-dihydropteroate synthase, ... | | Authors: | Chitnumsub, P, Jaruwat, A, Yuthavong, Y. | | Deposit date: | 2019-04-21 | | Release date: | 2020-02-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The structure of Plasmodium falciparum hydroxymethyldihydropterin pyrophosphokinase-dihydropteroate synthase reveals the basis of sulfa resistance.
Febs J., 287, 2020
|
|
8RIY
 
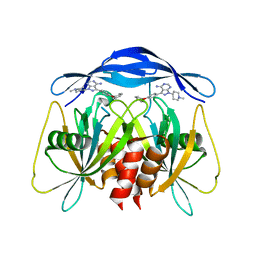 | | Human NUDT5 with ibrutinib derivative | | Descriptor: | 1-(1-methylpiperidin-4-yl)-3-(4-phenoxyphenyl)pyrazolo[3,4-d]pyrimidin-4-amine, ADP-sugar pyrophosphatase | | Authors: | Balikci-Akil, E, Elkins, J.M, Huber, K.V.M. | | Deposit date: | 2023-12-19 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.288 Å) | | Cite: | Unexpected Noncovalent Off-Target Activity of Clinical BTK Inhibitors Leads to Discovery of a Dual NUDT5/14 Antagonist.
J.Med.Chem., 67, 2024
|
|
8RDZ
 
 | |
6KLV
 
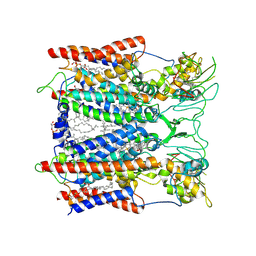 | | Hyperthermophilic respiratory Complex III | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, 2-[(2~{E},6~{E},10~{Z},14~{Z},18~{Z},23~{R})-3,7,11,15,19,23,27-heptamethyloctacosa-2,6,10,14,18-pentaenyl]naphthalene-1,4-dione, ANTIMYCIN, ... | | Authors: | Fei, S, Hartmut, M, Yun, Z, Guohong, P, Guoliang, Z, Hui, Z, Shuangbo, Z, Xiaoyun, P, Yan, Z. | | Deposit date: | 2019-07-30 | | Release date: | 2020-05-13 | | Last modified: | 2021-04-07 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A 3.3 angstrom -Resolution Structure of Hyperthermophilic Respiratory Complex III Reveals the Mechanism of Its Thermal Stability.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
4Q1C
 
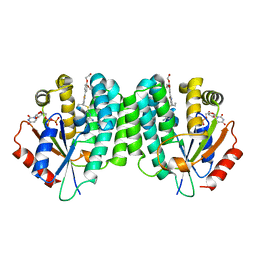 | | Human dCK C4S-S74E mutant in complex with UDP and the inhibitor 8 {2,2'-[{4-[(2R)-4-{[(4,6-diaminopyrimidin-2-yl)sulfanyl]methyl}-5-propyl-2,3-dihydro-1,3-thiazol-2-yl]benzene-1,2-diyl}bis(oxy)]diethanol} | | Descriptor: | 2,2'-((4-(4-(((4,6-diaminopyrimidin-2-yl)thio)methyl)-5-propylthiazol-2-yl)-1,2-phenylene)bis(oxy))bis(ethan-1-ol), Deoxycytidine kinase, URIDINE-5'-DIPHOSPHATE | | Authors: | Nomme, J, Lavie, A. | | Deposit date: | 2014-04-03 | | Release date: | 2014-11-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-guided development of deoxycytidine kinase inhibitors with nanomolar affinity and improved metabolic stability.
J.Med.Chem., 57, 2014
|
|
6KYK
 
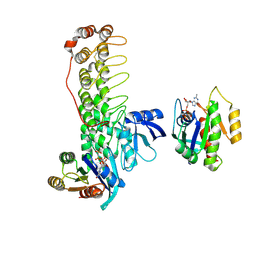 | | Crystal structure of Shank3 NTD-ANK mutant in complex with Rap1 | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ras-related protein Rap-1b, ... | | Authors: | Cai, Q, Zhang, M. | | Deposit date: | 2019-09-19 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Shank3 Binds to and Stabilizes the Active Form of Rap1 and HRas GTPases via Its NTD-ANK Tandem with Distinct Mechanisms.
Structure, 28, 2020
|
|
6KXG
 
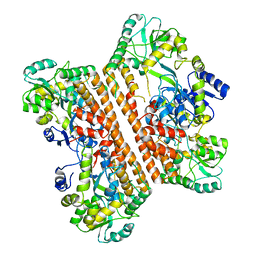 | | Crystal structure of caspase-11-CARD | | Descriptor: | caspase-11-CARD | | Authors: | Liu, M.Z.Y, Jin, T.C. | | Deposit date: | 2019-09-11 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.805 Å) | | Cite: | Crystal structure of caspase-11 CARD provides insights into caspase-11 activation.
Cell Discov, 6, 2020
|
|
6L1A
 
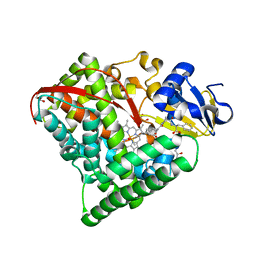 | | Crystal Structure of P450BM3 with N-enanthoyl-L-prolyl-L-phenylalanine | | Descriptor: | (2S)-2-[[(2S)-1-heptanoylpyrrolidin-2-yl]carbonylamino]-3-phenyl-propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, DIMETHYL SULFOXIDE, ... | | Authors: | Shoji, O, Yonemura, K. | | Deposit date: | 2019-09-28 | | Release date: | 2020-09-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Systematic Evolution of Decoy Molecules for the Highly Efficient Hydroxylation of Benzene and Small Alkanes Catalyzed by Wild-Type Cytochrome P450BM3
Acs Catalysis, 10, 2020
|
|
6KYH
 
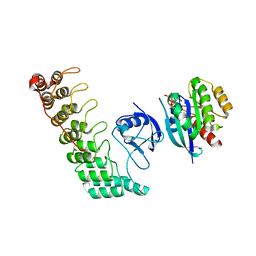 | | Crystal structure of Shank3 NTD-ANK A42K mutant in complex with HRas | | Descriptor: | GTPase HRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Cai, Q, Zhang, M. | | Deposit date: | 2019-09-18 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Shank3 Binds to and Stabilizes the Active Form of Rap1 and HRas GTPases via Its NTD-ANK Tandem with Distinct Mechanisms.
Structure, 28, 2020
|
|
5I56
 
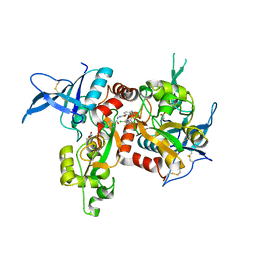 | | Agonist-bound GluN1/GluN2A agonist binding domains with TCN201 | | Descriptor: | GLUTAMIC ACID, GLYCINE, Glutamate receptor ionotropic, ... | | Authors: | Mou, T.-C, Sprang, S.R, Hansen, K.B. | | Deposit date: | 2016-02-14 | | Release date: | 2016-09-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural Basis for Negative Allosteric Modulation of GluN2A-Containing NMDA Receptors.
Neuron, 91, 2016
|
|
6L76
 
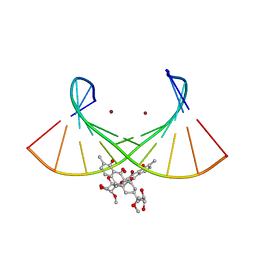 | | Crystal structure of the Ni(II)(Chro)2-d(TTGGGCCGAA/TTCGGCCCAA) complex at 2.94 angstrom resolution | | Descriptor: | (1S)-5-deoxy-1-O-methyl-1-C-[(2R,3S)-3,5,7,10-tetrahydroxy-6-methyl-4-oxo-1,2,3,4-tetrahydroanthracen-2-yl]-D-xylulose, 2,6-dideoxy-4-O-methyl-alpha-D-galactopyranose-(1-3)-(2R,3R,6R)-6-hydroxy-2-methyltetrahydro-2H-pyran-3-yl acetate, 3-C-methyl-4-O-acetyl-alpha-L-Olivopyranose-(1-3)-(2R,5S,6R)-6-methyltetrahydro-2H-pyran-2,5-diol-(1-3)-(2R,5S,6R)-6-methyltetrahydro-2H-pyran-2,5-diol, ... | | Authors: | Hou, M.H, Jhan, C.R, Satange, R.B, Lin, S.M. | | Deposit date: | 2019-10-31 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Targeting the ALS/FTD-associated A-DNA kink with anthracene-based metal complex causes DNA backbone straightening and groove contraction.
Nucleic Acids Res., 49, 2021
|
|
3IK0
 
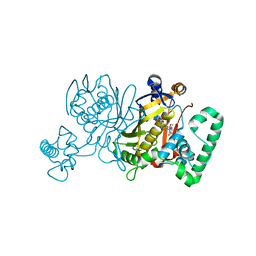 | | Lactobacillus casei Thymidylate Synthase in Ternary Complex with dUMP and the Phtalimidic Derivative 7C1 | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 4-(4-methyl-1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)benzenecarboximidamide, Thymidylate synthase | | Authors: | Pozzi, C, Cancian, L, Leone, R, Luciani, R, Ferrari, S, Mangani, S, Costi, M.P. | | Deposit date: | 2009-08-05 | | Release date: | 2010-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of the binding modes of N-phenylphthalimides inhibiting bacterial thymidylate synthase through X-ray crystallography screening
J.Med.Chem., 54, 2011
|
|
4H2M
 
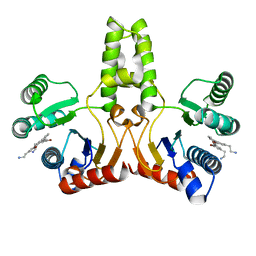 | | Structure of E. coli undecaprenyl diphosphate synthase in complex with BPH-1408 | | Descriptor: | 2,2'-{benzene-1,3-diylbis[ethyne-2,1-diyl(5-bromobenzene-3,1-diyl)]}diethanamine, Undecaprenyl pyrophosphate synthase | | Authors: | Zhu, W, Oldfield, E. | | Deposit date: | 2012-09-12 | | Release date: | 2012-12-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Antibacterial drug leads targeting isoprenoid biosynthesis.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6O1H
 
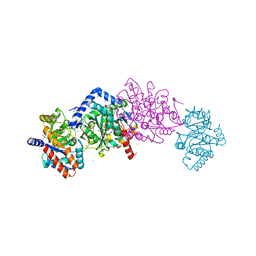 | | Tryptophan synthase Q114A mutant in complex with N-(4'-trifluoromethoxybenzenesulfonyl)-2-amino-1-ethylphosphate (F9F) at the enzyme alpha-site, cesium ion at the metal coordination site, and 2-aminophenol quinonoid at the enzyme beta site | | Descriptor: | (2E)-2-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)imino]-3-[(2-hydroxyphenyl)amino]propanoic acid, 2-({[4-(TRIFLUOROMETHOXY)PHENYL]SULFONYL}AMINO)ETHYL DIHYDROGEN PHOSPHATE, CESIUM ION, ... | | Authors: | Hilario, E, Dunn, M.F, Mueller, L.J, Fan, L. | | Deposit date: | 2019-02-19 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal structure of Salmonella typhimurium Tryptophan Synthase mutant beta-Q114A with 2-({[4-(trifluoromethoxy)phenyl]sulfonyl}amino)ethyl dihydrogen phosphate (F9F) at the alpha-site, Cesium ion at the metal coordination site, and [3-hydroxy-2-methyl-5-phosphonooxymethyl-pyridin-4-ylmethyl]-serine (PLS) at the beta-site.
To be Published
|
|
1AMN
 
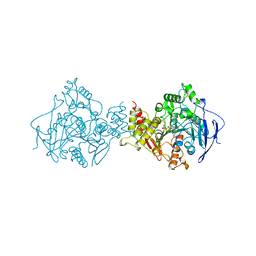 | | TRANSITION STATE ANALOG: ACETYLCHOLINESTERASE COMPLEXED WITH M-(N,N,N-TRIMETHYLAMMONIO)TRIFLUOROACETOPHENONE | | Descriptor: | ACETYLCHOLINESTERASE, M-(N,N,N-TRIMETHYLAMMONIO)-2,2,2-TRIFLUORO-1,1-DIHYDROXYETHYLBENZENE, SULFATE ION | | Authors: | Harel, M, Silman, I, Sussman, J.L. | | Deposit date: | 1996-02-13 | | Release date: | 1996-04-03 | | Last modified: | 2021-06-02 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The X-ray structure of a transition state analog complex reveals the molecular origins of the catalytic power and substrate specificity of acetylcholinesterase.
J.Am.Chem.Soc., 118, 1996
|
|
5K58
 
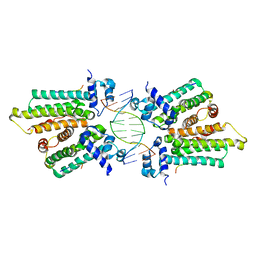 | |
6QEI
 
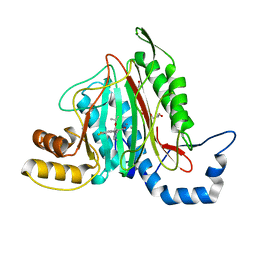 | | CRYSTAL STRUCTURE OF HUMAN METHIONINE AMINOPEPTIDASE-2 IN COMPLEX WITH AN INHIBITOR 5,6-Difluoro-3-(2-isopropoxy-4-piperazin-1-yl-phenyl)-1H-indole-2-carboxylic acid amide | | Descriptor: | 1,2-ETHANEDIOL, 5,6-bis(fluoranyl)-3-(4-piperazin-1-yl-2-propan-2-yloxy-phenyl)-1~{H}-indole-2-carboxamide, DIMETHYL SULFOXIDE, ... | | Authors: | Musil, D, Heinrich, T, Lehmann, M. | | Deposit date: | 2019-01-07 | | Release date: | 2019-05-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery and Structure-Based Optimization of Next-Generation Reversible Methionine Aminopeptidase-2 (MetAP-2) Inhibitors.
J.Med.Chem., 62, 2019
|
|
7XQE
 
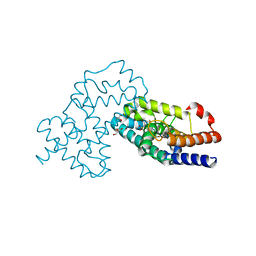 | | Crystal Structure of human RORgamma (C455E) LBD in complex with compound XY039 | | Descriptor: | 2,4-difluoro-N-(1-((4-(trifluoromethyl)benzyl)sulfonyl)-1,2,3,4-tetrahydroquinolin-7-yl)benzenesulfonamide, ETHANOL, GLYCEROL, ... | | Authors: | Wu, X, Li, C, Zhang, Y, Xu, Y. | | Deposit date: | 2022-05-07 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Discovery and pharmacological characterization of 1,2,3,4-tetrahydroquinoline derivatives as ROR gamma inverse agonists against prostate cancer.
Acta Pharmacol.Sin., 2024
|
|
6QFV
 
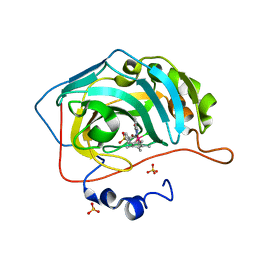 | | Human carbonic anhydrase II with bound IrCp* complex (cofactor 8) to generate an artificial transfer hydrogenase (ATHase) | | Descriptor: | 4-[2-(9-chloranyl-2',3',4',5',6'-pentamethyl-7-oxidanylidene-spiro[1$l^{4},8-diaza-9$l^{8}-iridabicyclo[4.3.0]nona-1(6),2,4-triene-9,1'-1$l^{8}-iridapentacyclo[2.2.0.0^{1,3}.0^{1,5}.0^{2,6}]hexane]-8-yl)ethyl]benzenesulfonamide, Carbonic anhydrase 2, SULFATE ION, ... | | Authors: | Rebelein, J.G. | | Deposit date: | 2019-01-10 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Chemical Optimization of Whole-Cell Transfer Hydrogenation Using Carbonic Anhydrase as Host Protein.
Acs Catalysis, 9, 2019
|
|
7X2Z
 
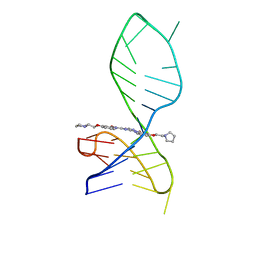 | | NMR solution structure of the 1:1 complex of a pyridostatin derivative (PyPDS) bound to a G-quadruplex MYT1L | | Descriptor: | 4-(2-azanylethoxy)-N2,N6-bis[4-(2-pyrrolidin-1-ylethoxy)quinolin-2-yl]pyridine-2,6-dicarboxamide, G-quadruplex DNA MYT1L | | Authors: | Liu, L.-Y, Mao, Z.-W, Liu, W. | | Deposit date: | 2022-02-26 | | Release date: | 2022-06-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of Pyridostatin and Its Derivatives Specifically Binding to G-Quadruplexes.
J.Am.Chem.Soc., 144, 2022
|
|
