7LNL
 
 | | Crystal structure of KPC-2 S70G/T215P mutant with hydrolyzed imipenem | | Descriptor: | (2R)-2-[(2S,3R)-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-4-(2-methanimidamidoethylsulfanyl)-2,3-dihydro-1H-pyrrole -5-carboxylic acid, Carbapenem-hydrolyzing beta-lactamase KPC | | Authors: | Furey, I, Palzkill, T, Sankaran, B, Hu, L, Prasad, B.V.V. | | Deposit date: | 2021-02-07 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Local interactions with the Glu166 base and the conformation of an active site loop play key roles in carbapenem hydrolysis by the KPC-2 beta-lactamase.
J.Biol.Chem., 296, 2021
|
|
2VCT
 
 | |
7LHE
 
 | | Structure of full-length IP3R1 channel reconstituted into lipid nanodisc in the apo-state | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, Inositol 1,4,5-trisphosphate receptor type 1, ZINC ION | | Authors: | Baker, M.R, Fan, G, Baker, M.L, Serysheva, I.I. | | Deposit date: | 2021-01-22 | | Release date: | 2021-06-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of type 1 IP3R channel in a lipid bilayer
Commun Biol, 4, 2021
|
|
7LPS
 
 | | Crystal structure of DDB1-CRBN-ALV1 complex bound to Helios (IKZF2 ZF2) | | Descriptor: | 3-[3-[[1-[(3~{S})-2,6-bis(oxidanylidene)piperidin-3-yl]-2,5-bis(oxidanylidene)pyrrol-3-yl]amino]phenyl]-~{N}-(3-chloranyl-4-methyl-phenyl)propanamide, DNA damage-binding protein 1, Protein cereblon, ... | | Authors: | Nowak, R.P, Fischer, E.S. | | Deposit date: | 2021-02-12 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.78 Å) | | Cite: | Acute pharmacological degradation of Helios destabilizes regulatory T cells.
Nat.Chem.Biol., 17, 2021
|
|
8UXX
 
 | | Arp2/3 branch junction complex, BeFx state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Chavali, S.S, Chou, S.Z, Sindelar, C.V. | | Deposit date: | 2023-11-11 | | Release date: | 2024-01-31 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures reveal how phosphate release from Arp3 weakens actin filament branches formed by Arp2/3 complex.
Nat Commun, 15, 2024
|
|
1O6V
 
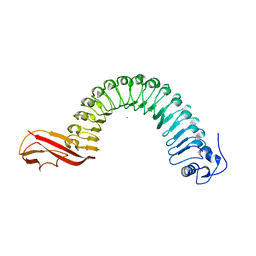 | | Internalin (INLA, Listeria monocytogenes) - functional domain, uncomplexed | | Descriptor: | CALCIUM ION, INTERNALIN A | | Authors: | Schubert, W.-D, Urbanke, C, Ziehm, T, Beier, V, Machner, M.P, Domann, E, Wehland, J, Chakraborty, T, Heinz, D.W. | | Deposit date: | 2002-10-16 | | Release date: | 2002-12-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of Internalin, a Major Invasion Protein of Listeria Monocytogenes, in Complex with its Human Receptor E-Cadherin
Cell(Cambridge,Mass.), 111, 2002
|
|
7LHF
 
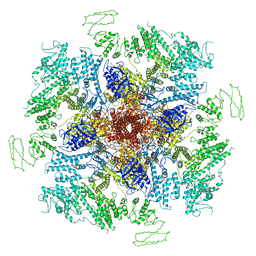 | | Structure of full-length IP3R1 channel solubilized in LNMG & lipid in the apo-state | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, Inositol 1,4,5-trisphosphate receptor type 1, ZINC ION | | Authors: | Baker, M.R, Fan, G, Baker, M.L, Serysheva, I.I. | | Deposit date: | 2021-01-22 | | Release date: | 2021-06-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Cryo-EM structure of type 1 IP3R channel in a lipid bilayer
Commun Biol, 4, 2021
|
|
5EIR
 
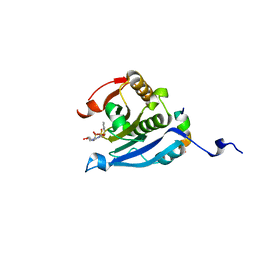 | | Co-crystal structure of eIF4E with nucleotide mimetic inhibitor. | | Descriptor: | Eukaryotic translation initiation factor 4 gamma 1, Eukaryotic translation initiation factor 4E, SULFATE ION, ... | | Authors: | Nowicki, M.W, Walkinshaw, M.D, Fischer, P.M. | | Deposit date: | 2015-10-30 | | Release date: | 2016-09-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Design of nucleotide-mimetic and non-nucleotide inhibitors of the translation initiation factor eIF4E: Synthesis, structural and functional characterisation.
Eur.J.Med.Chem., 124, 2016
|
|
8U7V
 
 | | Human retinal variant phosphomimetic IMPDH1(546)-S477D filament bound by GTP, ATP, IMP, and NAD+, interface-centered | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, INOSINIC ACID, ... | | Authors: | Calise, S.J, Kollman, J.M. | | Deposit date: | 2023-09-15 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Light-sensitive phosphorylation regulates retinal IMPDH1 activity and filament assembly.
J.Cell Biol., 223, 2024
|
|
2VLK
 
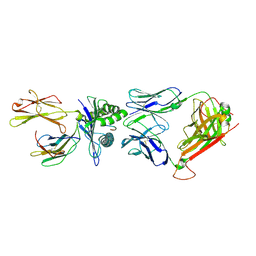 | | The Structural Dynamics and Energetics of an Immunodominant T-cell Receptor are Programmed by its Vbeta Domain | | Descriptor: | BETA-2-MICROGLOBULIN, FLU MATRIX PEPTIDE, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, ... | | Authors: | Ishizuka, J, Stewart-Jones, G, Van Der Merwe, A, Bell, J, Mcmichael, A, Jones, Y. | | Deposit date: | 2008-01-15 | | Release date: | 2008-01-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Structural Dynamics and Energetics of an Immunodominant T-Cell Receptor are Programmed by its Vbeta Domain
Immunity, 28, 2008
|
|
1ODS
 
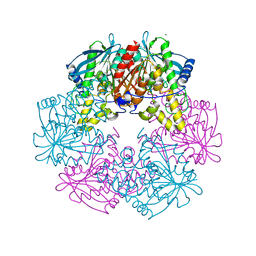 | | Cephalosporin C deacetylase from Bacillus subtilis | | Descriptor: | CEPHALOSPORIN C DEACETYLASE, CHLORIDE ION, MAGNESIUM ION | | Authors: | Vincent, F, Charnock, S.J, Verschueren, K.H.G, Turkenburg, J.P, Scott, D.J, Offen, W.A, Roberts, S, Pell, G, Gilbert, H.J, Brannigan, J.A, Davies, G.J. | | Deposit date: | 2003-02-20 | | Release date: | 2003-07-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Multifunctional Xylooligosaccharide/Cephalosporin C Deacetylase Revealed by the Hexameric Structure of the Bacillus Subtilis Enzyme at 1.9A Resolution
J.Mol.Biol., 330, 2003
|
|
4NAA
 
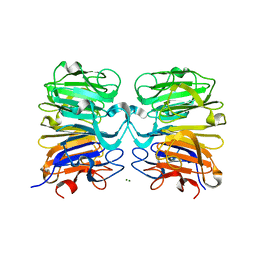 | | Crystal structure of UVB photoreceptor UVR8 from Arabidopsis thaliana and UV-induced structural changes at 120K | | Descriptor: | MAGNESIUM ION, Ultraviolet-B receptor UVR8 | | Authors: | Yang, X, Zeng, X, Ren, Z, Zhao, K.H. | | Deposit date: | 2013-10-22 | | Release date: | 2016-10-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Dynamic Crystallography Reveals Early Signalling Events in Ultraviolet Photoreceptor UVR8.
Nat Plants, 1, 2015
|
|
4NBA
 
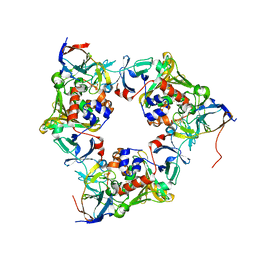 | | Carbazole-bound oxygenase with Ile262 replaced by Val and ferredoxin complex of carbazole 1,9a-dioxygenase | | Descriptor: | 9H-CARBAZOLE, FE (II) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Ashikawa, Y, Usami, Y, Inoue, K, Nojiri, H. | | Deposit date: | 2013-10-23 | | Release date: | 2014-03-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of the divergent oxygenation reactions catalyzed by the rieske nonheme iron oxygenase carbazole 1,9a-dioxygenase.
Appl.Environ.Microbiol., 80, 2014
|
|
4IN4
 
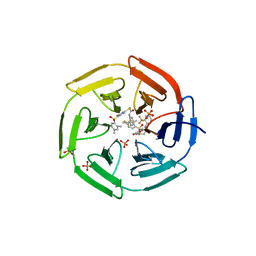 | | Crystal structure of cpd 15 bound to Keap1 Kelch domain | | Descriptor: | 2-({5-[(2,4-dimethylphenyl)sulfonyl]-6-oxo-1,6-dihydropyrimidin-2-yl}sulfanyl)-N-[2-(trifluoromethyl)phenyl]acetamide, Kelch-like ECH-associated protein 1, PHOSPHATE ION | | Authors: | Silvian, L, Marcotte, D. | | Deposit date: | 2013-01-03 | | Release date: | 2013-05-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Small molecules inhibit the interaction of Nrf2 and the Keap1 Kelch domain through a non-covalent mechanism.
Bioorg.Med.Chem., 21, 2013
|
|
4NBS
 
 | | The structure of aIF2gamma subunit H20F from archaeon Sulfolobus solfataricus complexed with GDPCP | | Descriptor: | FORMIC ACID, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, ... | | Authors: | Kravchenko, O.V, Nikonov, O.S, Arhipova, V.I, Stolboushkina, E.A, Gabdulkhakov, A.G, Nikulin, A.D, Garber, M.B, Nikonov, S.V. | | Deposit date: | 2013-10-23 | | Release date: | 2014-10-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.305 Å) | | Cite: | The structure of aIF2gamma subunit H20F from archaeon Sulfolobus solfataricus complexed with GDPCP
To be Published
|
|
5ENL
 
 | |
3G5H
 
 | | Crystallographic analysis of cytochrome P450 cyp121 | | Descriptor: | (3S,6S)-3,6-bis(4-hydroxybenzyl)piperazine-2,5-dione, Cytochrome P450 121, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Belin, P, Le Du, M.H, Gondry, M. | | Deposit date: | 2009-02-05 | | Release date: | 2009-04-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Identification and structural basis of the reaction catalyzed by CYP121, an essential cytochrome P450 in Mycobacterium tuberculosis.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
8U7M
 
 | | Human retinal variant phosphomimetic IMPDH1(595)-S477D free octamer bound by GTP, ATP, IMP, and NAD+ | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, INOSINIC ACID, ... | | Authors: | Calise, S.J, Kollman, J.M. | | Deposit date: | 2023-09-15 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Light-sensitive phosphorylation regulates retinal IMPDH1 activity and filament assembly.
J.Cell Biol., 223, 2024
|
|
4MTW
 
 | | Thermolysin in complex with UBTLN36 | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Krimmer, S.G, Heine, A, Klebe, G. | | Deposit date: | 2013-09-20 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Methyl, Ethyl, Propyl, Butyl: Futile But Not for Water, as the Correlation of Structure and Thermodynamic Signature Shows in a Congeneric Series of Thermolysin Inhibitors.
Chemmedchem, 4, 2014
|
|
2VBM
 
 | | Tailspike protein of bacteriophage Sf6 complexed with tetrasaccharide | | Descriptor: | MAGNESIUM ION, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Mueller, J.J, Barbirz, S, Freiberg, A, Seckler, R, Heinemann, U. | | Deposit date: | 2007-09-14 | | Release date: | 2008-04-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An Intersubunit Active Site between Supercoiled Parallel Beta Helices in the Trimeric Tailspike Endorhamnosidase of Shigella Flexneri Phage Sf6.
Structure, 16, 2008
|
|
2V2T
 
 | | X-ray structure of a NF-kB p50-RelB-DNA complex | | Descriptor: | 5'-D(*CP*GP*GP*GP*AP*AP*TP*TP*CP*CP*CP)-3', NUCLEAR FACTOR NF-KAPPA-B P105 SUBUNIT, TRANSCRIPTION FACTOR RELB | | Authors: | Moorthy, A.K, Huang, D.B, Wang, V.Y, Vu, D, Ghosh, G. | | Deposit date: | 2007-06-07 | | Release date: | 2007-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | X-Ray Structure of a NF-kappaB p50/Relb/DNA Complex Reveals Assembly of Multiple Dimers on Tandem kappaB Sites.
J.Mol.Biol., 373, 2007
|
|
7RF9
 
 | | O2-, PLP-dependent desaturase Plu4 intermediate-bound enzyme | | Descriptor: | (2E)-5-carbamimidamido-2-{[(Z)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4(1H)-ylidene}methyl]imino}pentanoic acid, 1,2-ETHANEDIOL, 2-(2-ETHOXYETHOXY)ETHANOL, ... | | Authors: | Hoffarth, E.R, Ryan, K.S. | | Deposit date: | 2021-07-13 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.926 Å) | | Cite: | A shared mechanistic pathway for pyridoxal phosphate-dependent arginine oxidases.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
2V6L
 
 | | Molecular Model of a Type III Secretion System Needle | | Descriptor: | MXIH | | Authors: | Deane, J.E, Roversi, P, Cordes, F.S, Johnson, S, Kenjale, R, Daniell, S, Booy, F, Picking, W.L, Picking, W.D, Blocker, A.J, Lea, S.M. | | Deposit date: | 2007-07-19 | | Release date: | 2007-07-31 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (16 Å) | | Cite: | Molecular model of a type III secretion system needle: Implications for host-cell sensing.
Proc. Natl. Acad. Sci. U.S.A., 103, 2006
|
|
4IDD
 
 | | Structure of the Fragaria x ananassa enone oxidoreductase in complex with NADPH and EHMF | | Descriptor: | (2R)-2-ethyl-4-hydroxy-5-methylfuran-3(2H)-one, 1,2-ETHANEDIOL, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Schiefner, A, Skerra, A. | | Deposit date: | 2012-12-12 | | Release date: | 2013-04-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for the enzymatic formation of the key strawberry flavor compound 4-hydroxy-2,5-dimethyl-3(2H)-furanone
J.Biol.Chem., 288, 2013
|
|
3FXT
 
 | | Crystal structure of the N-terminal domain of human NUDT6 | | Descriptor: | GLYCEROL, Nucleoside diphosphate-linked moiety X motif 6 | | Authors: | Tresaugues, L, Welin, M, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Karlberg, T, Kotenyova, T, Lehtio, L, Moche, M, Nilsson, M.E, Nyman, T, Persson, C, Sagemark, J, Schueler, H, Siponen, M.I, Thorsell, A.G, Van Den Berg, S, Weigelt, J, Wikstrom, M, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-01-21 | | Release date: | 2009-03-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the N-terminal domain of human NUDT6
To be Published
|
|
