4X1E
 
 | | Crystal structure of unliganded E. coli transcriptional regulator RutR, W167A mutant | | Descriptor: | HTH-type transcriptional regulator RutR | | Authors: | Nguyen Le Minh, P, de Cima, S, Bervoets, I, Maes, D, Rubio, V, Charlier, D. | | Deposit date: | 2014-11-24 | | Release date: | 2015-01-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Ligand binding specificity of RutR, a member of the TetR family of transcription regulators in Escherichia coli.
Febs Open Bio, 5, 2015
|
|
4XR5
 
 | | X-ray structure of the unliganded thymidine phosphorylase from Salmonella typhimurium at 2.05 A resolution | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, TRIETHYLENE GLYCOL, ... | | Authors: | Balaev, V.V, Lashkov, A.A, Gabdulkhakov, A.G, Betzel, C, Mikhailov, A.M. | | Deposit date: | 2015-01-20 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural investigation of the thymidine phosphorylase from Salmonella typhimurium in the unliganded state and its complexes with thymidine and uridine.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
8TVP
 
 | | Cryo-EM structure of CPD-stalled Pol II in complex with Rad26 (open state) | | Descriptor: | DNA (NTS), DNA (TS), DNA repair and recombination protein RAD26, ... | | Authors: | Sarsam, R.D, Lahiri, I, Leschziner, A.E. | | Deposit date: | 2023-08-18 | | Release date: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Elf1 promotes Rad26's interaction with lesion-arrested Pol II for transcription-coupled repair.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8TVQ
 
 | | Cryo-EM structure of CPD stalled 10-subunit Pol II in complex with Rad26 | | Descriptor: | DNA (NTS), DNA (TS), DNA repair and recombination protein RAD26, ... | | Authors: | Sarsam, R.D, Lahiri, I, Leschziner, A.E. | | Deposit date: | 2023-08-18 | | Release date: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Elf1 promotes Rad26's interaction with lesion-arrested Pol II for transcription-coupled repair.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
4Y7N
 
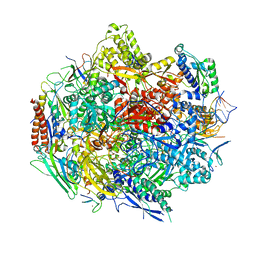 | | The Structure Insight into 5-Carboxycytosine Recognition by RNA Polymerase II during Transcription Elongation. | | Descriptor: | DNA (29-MER), DNA (5'-D(*CP*TP*GP*CP*TP*TP*AP*TP*CP*GP*GP*TP*AP*G)-3'), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Wang, L, Chong, J, Wang, D. | | Deposit date: | 2015-02-15 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Molecular basis for 5-carboxycytosine recognition by RNA polymerase II elongation complex.
Nature, 523, 2015
|
|
8TVX
 
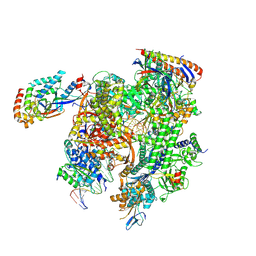 | | Cryo-EM structure of CPD-stalled Pol II (Conformation 2) | | Descriptor: | DNA (NTS), DNA (TS), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Sarsam, R.D, Lahiri, I, Leschziner, A.E. | | Deposit date: | 2023-08-18 | | Release date: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Elf1 promotes Rad26's interaction with lesion-arrested Pol II for transcription-coupled repair.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8TVY
 
 | | Cryo-EM structure of CPD lesion containing RNA Polymerase II elongation complex with Rad26 and Elf1 (closed state) | | Descriptor: | DNA (NTS), DNA (TS), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Sarsam, R.D, Lahiri, I, Leschziner, A.E. | | Deposit date: | 2023-08-18 | | Release date: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Elf1 promotes Rad26's interaction with lesion-arrested Pol II for transcription-coupled repair.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8TVW
 
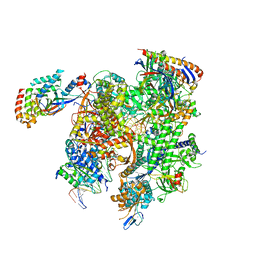 | | Cryo-EM structure of CPD-stalled Pol II (conformation 1) | | Descriptor: | DNA (NTS), DNA (TS), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Sarsam, R.D, Lahiri, I, Leschziner, A.E. | | Deposit date: | 2023-08-18 | | Release date: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Elf1 promotes Rad26's interaction with lesion-arrested Pol II for transcription-coupled repair.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8TUG
 
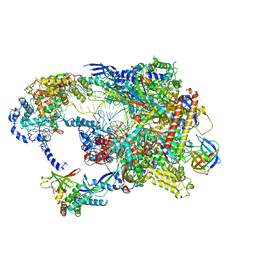 | | Cryo-EM structure of CPD-stalled Pol II in complex with Rad26 (engaged state) | | Descriptor: | DNA (NTS), DNA (TS), DNA repair and recombination protein RAD26, ... | | Authors: | Sarsam, R.D, Lahiri, I, Leschziner, A.E. | | Deposit date: | 2023-08-16 | | Release date: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Elf1 promotes Rad26's interaction with lesion-arrested Pol II for transcription-coupled repair.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
4Y52
 
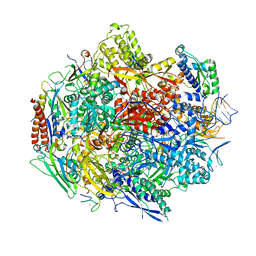 | | Crystal structure of 5-Carboxycytosine Recognition by RNA Polymerase II during Transcription Elongation. | | Descriptor: | DNA (29-MER), DNA (5'-D(*CP*TP*GP*CP*TP*TP*AP*TP*CP*GP*GP*TP*AP*G)-3'), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Wang, L, Chong, J, Wang, D. | | Deposit date: | 2015-02-11 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Molecular basis for 5-carboxycytosine recognition by RNA polymerase II elongation complex.
Nature, 523, 2015
|
|
4YEK
 
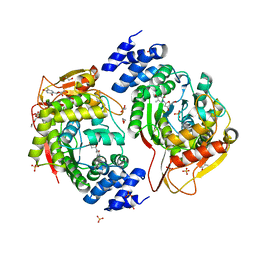 | | X-ray structure of the thymidine phosphorylase from Salmonella typhimurium in complex with thymidine | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, SULFATE ION, ... | | Authors: | Balaev, V.V, Lashkov, A.A, Gabdulkhakov, A.G, Betzel, C, Mikhailov, A.M. | | Deposit date: | 2015-02-24 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural investigation of the thymidine phosphorylase from Salmonella typhimurium in the unliganded state and its complexes with thymidine and uridine.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
1NGT
 
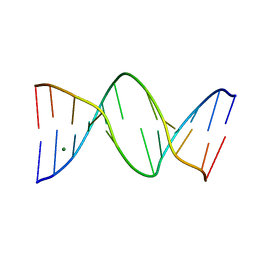 | | The Role of Minor Groove Functional Groups in DNA Hydration | | Descriptor: | 5'-D(*CP*GP*CP*GP*AP*AP*(MTR)P*TP*CP*GP*CP*G)-3', MAGNESIUM ION | | Authors: | Woods, K.K, Lan, T, McLaughlin, L.W, Williams, L.D. | | Deposit date: | 2002-12-17 | | Release date: | 2003-03-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | The Role of Minor Groove Functional Groups in DNA Hydration
Nucleic Acids Res., 31, 2003
|
|
7DFY
 
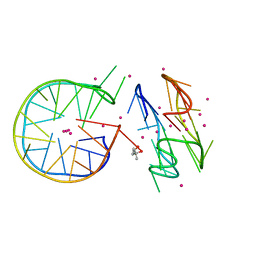 | | Novel motif for left-handed G-quadruplex formation | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2xMotif2, POTASSIUM ION, ... | | Authors: | Das, P, Winnerdy, F.R, Maity, A, Mechulam, Y, Phan, A.T. | | Deposit date: | 2020-11-10 | | Release date: | 2021-09-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | A novel minimal motif for left-handed G-quadruplex formation.
Chem.Commun.(Camb.), 57, 2021
|
|
6OD5
 
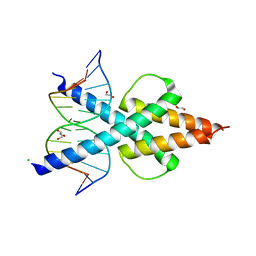 | | Human TCF4 C-terminal bHLH domain in Complex with 12-bp Oligonucleotide Containing E-box Sequence with 5-carboxylcytosines | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA (5'-D(*AP*(1CC)P*GP*CP*AP*CP*GP*TP*GP*(1CC)P*G)-3'), ... | | Authors: | Horton, J.R, Cheng, X, Yang, J. | | Deposit date: | 2019-03-25 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for preferential binding of human TCF4 to DNA containing 5-carboxylcytosine.
Nucleic Acids Res., 47, 2019
|
|
3Q61
 
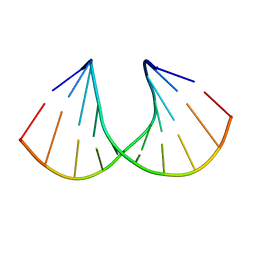 | | 3'-Fluoro Hexitol Nucleic Acid DNA Structure | | Descriptor: | DNA (5'-D(*GP*CP*GP*TP*AP*(F3H)P*AP*CP*GP*C)-3') | | Authors: | Seth, P.R, Allerson, C.R, Prakash, T.P, Siwkowski, A, Berdeja, A, Yu, J, Pallan, P.S, Watt, A.T, Gaus, H, Bhat, B, Egli, M, Swayze, E.E. | | Deposit date: | 2010-12-30 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Synthesis, improved antisense activity and structural rationale for the divergent RNA affinities of 3'-fluoro hexitol nucleic acid (FHNA and Ara-FHNA) modified oligonucleotides.
J.Am.Chem.Soc., 133, 2011
|
|
5ITT
 
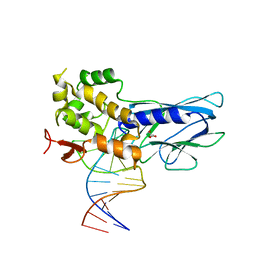 | | Crystal Structure of Human NEIL1 bound to duplex DNA containing THF | | Descriptor: | DNA (26-MER), Endonuclease 8-like 1, GLYCEROL | | Authors: | Zhu, C, Lu, L, Zhang, J, Yue, Z, Song, J, Zong, S, Liu, M, Stovicek, O, Gao, Y, Yi, C. | | Deposit date: | 2016-03-17 | | Release date: | 2016-07-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Tautomerization-dependent recognition and excision of oxidation damage in base-excision DNA repair
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5ITR
 
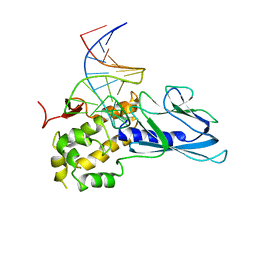 | | Crystal Structure of Human NEIL1(P2G) bound to duplex DNA containing THF | | Descriptor: | DNA (26-MER), Endonuclease 8-like 1 | | Authors: | Zhu, C, Lu, L, Zhang, J, Yue, Z, Song, J, Zong, S, Liu, M, Stovicek, O, Gao, Y, Yi, C. | | Deposit date: | 2016-03-17 | | Release date: | 2016-07-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Tautomerization-dependent recognition and excision of oxidation damage in base-excision DNA repair
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
2HBN
 
 | |
2V8G
 
 | | Crystal structure of beta-alanine synthase from Saccharomyces kluyveri in complex with the product beta-alanine | | Descriptor: | BETA-ALANINE, BETA-ALANINE SYNTHASE, BICINE, ... | | Authors: | Lundgren, S, Andersen, B, Piskur, J, Dobritzsch, D. | | Deposit date: | 2007-08-07 | | Release date: | 2007-10-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structures of Yeast -Alanine Synthase Complexes Reveal the Mode of Substrate Binding and Large Scale Domain Closure Movements.
J.Biol.Chem., 282, 2007
|
|
2V8V
 
 | | Crystal structure of mutant R322A of beta-alanine synthase from Saccharomyces kluyveri | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, BETA-ALANINE SYNTHASE, N-(AMINOCARBONYL)-BETA-ALANINE, ... | | Authors: | Lundgren, S, Andersen, B, Piskur, J, Dobritzsch, D. | | Deposit date: | 2007-08-15 | | Release date: | 2007-10-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structures of Yeast -Alanine Synthase Complexes Reveal the Mode of Substrate Binding and Large Scale Domain Closure Movements.
J.Biol.Chem., 282, 2007
|
|
2V8D
 
 | | Crystal structure of mutant E159A of beta-alanine synthase from Saccharomyces kluyveri | | Descriptor: | BETA-ALANINE SYNTHASE, ZINC ION | | Authors: | Lundgren, S, Andersen, B, Piskur, J, Dobritzsch, D. | | Deposit date: | 2007-08-07 | | Release date: | 2007-10-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of Yeast -Alanine Synthase Complexes Reveal the Mode of Substrate Binding and Large Scale Domain Closure Movements.
J.Biol.Chem., 282, 2007
|
|
2V8H
 
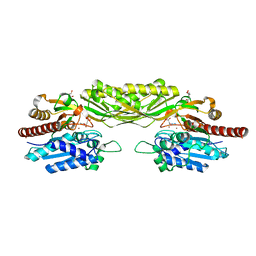 | | Crystal structure of mutant E159A of beta-alanine synthase from Saccharomyces kluyveri in complex with its substrate N-carbamyl-beta- alanine | | Descriptor: | BETA-ALANINE SYNTHASE, BICINE, N-(AMINOCARBONYL)-BETA-ALANINE, ... | | Authors: | Lundgren, S, Andersen, B, Piskur, J, Dobritzsch, D. | | Deposit date: | 2007-08-08 | | Release date: | 2007-10-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Yeast -Alanine Synthase Complexes Reveal the Mode of Substrate Binding and Large Scale Domain Closure Movements.
J.Biol.Chem., 282, 2007
|
|
6BNA
 
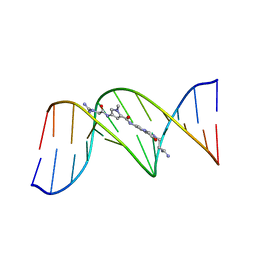 | | BINDING OF AN ANTITUMOR DRUG TO DNA. NETROPSIN AND C-G-C-G-A-A-T-T-BRC-G-C-G | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*(CBR)P*GP*CP*G)-3'), NETROPSIN | | Authors: | Kopka, M.L, Yoon, C, Goodsell, D, Pjura, P, Dickerson, R.E. | | Deposit date: | 1984-08-30 | | Release date: | 1984-10-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Binding of an antitumor drug to DNA, Netropsin and C-G-C-G-A-A-T-T-BrC-G-C-G.
J.Mol.Biol., 183, 1985
|
|
4YYY
 
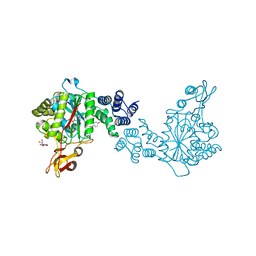 | | X-ray structure of the thymidine phosphorylase from Salmonella typhimurium in complex with uridine | | Descriptor: | CITRIC ACID, TRIETHYLENE GLYCOL, Thymidine phosphorylase, ... | | Authors: | Balaev, V.V, Lashkov, A.A, Gabdulkhakov, A.G, Betzel, C, Mikhailov, A.M. | | Deposit date: | 2015-03-24 | | Release date: | 2016-03-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural investigation of the thymidine phosphorylase from Salmonella typhimurium in the unliganded state and its complexes with thymidine and uridine.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
213D
 
 | |
