7PC2
 
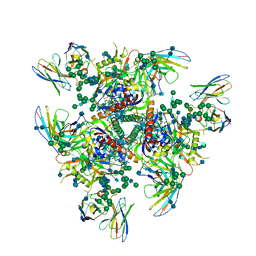 | | HIV-1 Env (BG505 SOSIP.664) in complex with the IgA bNAb 7-269 and the antibody 3BNC117. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3BNC IgG Fab heavy chain, ... | | Authors: | Fernandez, I, Bontems, F, Pehau-Arnaudet, G, Rey, F. | | Deposit date: | 2021-08-03 | | Release date: | 2022-02-23 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Epitope convergence of broadly HIV-1 neutralizing IgA and IgG antibody lineages in a viremic controller.
J.Exp.Med., 219, 2022
|
|
6USV
 
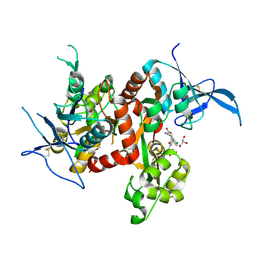 | | Crystal structure of GluN1/GluN2A ligand-binding domain in complex with glycine and SDZ 220-040 | | Descriptor: | (2S)-2-amino-3-[2',4'-dichloro-4-hydroxy-5-(phosphonomethyl)biphenyl-3-yl]propanoic acid, GLYCEROL, GLYCINE, ... | | Authors: | Romero-Hernandez, A, Tajima, N, Chou, T, Furukawa, h. | | Deposit date: | 2019-10-28 | | Release date: | 2020-07-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.304 Å) | | Cite: | Structural Basis of Functional Transitions in Mammalian NMDA Receptors.
Cell, 182, 2020
|
|
6USU
 
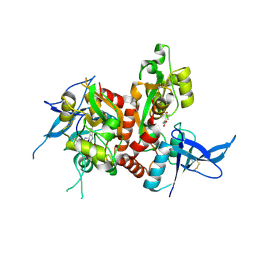 | | Crystal structure of GluN1/GluN2A ligand-binding domain in complex with L689,560 and glutamate | | Descriptor: | (2R,4S)-5,7-dichloro-4-[(phenylcarbamoyl)amino]-1,2,3,4-tetrahydroquinoline-2-carboxylic acid, GLUTAMIC ACID, Glutamate receptor ionotropic, ... | | Authors: | Romero-Hernandez, A, Tajima, N, Chou, T, Furukawa, H. | | Deposit date: | 2019-10-28 | | Release date: | 2020-07-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.092 Å) | | Cite: | Structural Basis of Functional Transitions in Mammalian NMDA Receptors.
Cell, 182, 2020
|
|
6WBO
 
 | |
3ZH3
 
 | |
8EYM
 
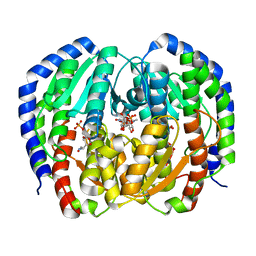 | | CRYSTAL STRUCTURE OF NAGB-II PHOSPHOSUGAR ISOMERASE FROM SHEWANELLA DENITRIFICANS OS217 IN COMPLEX WITH GLUCITOLAMINE-6-PHOSPHATE AND N-ACETYLGLUCOSAMINE-6-PHOSPHATE AT 2.31 A RESOLUTION | | Descriptor: | 2-DEOXY-2-AMINO GLUCITOL-6-PHOSPHATE, 2-acetamido-2-deoxy-6-O-phosphono-alpha-D-glucopyranose, GLUCOSAMINE-6-PHOSPHATE DEAMINASE | | Authors: | Rodriguez-Hernandez, A, Marcos-Viquez, J, Rodriguez-Romero, A, Bustos-Jaimes, I. | | Deposit date: | 2022-10-27 | | Release date: | 2023-05-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.311 Å) | | Cite: | Substrate binding in the allosteric site mimics homotropic cooperativity in the SIS-fold glucosamine-6-phosphate deaminases.
Protein Sci., 32, 2023
|
|
8EOL
 
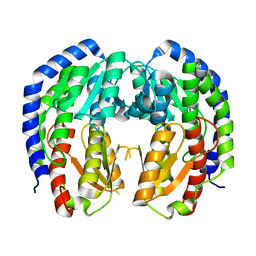 | |
1S4A
 
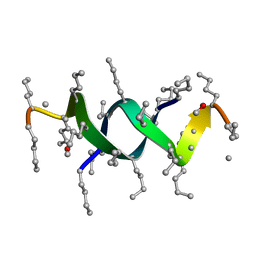 | |
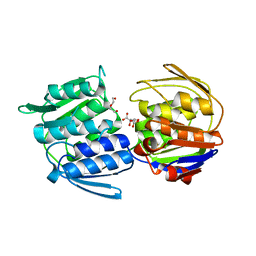
3ZH4
 
 | |
6C8Z
 
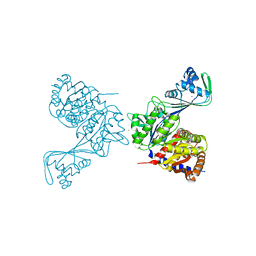 | | Last common ancestor of ADP-dependent phosphofructokinases from Methanosarcinales | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADP-dependent phosphofructokinase, MAGNESIUM ION, ... | | Authors: | Castro-Fernandez, V, Gonzalez-Ordenes, F, Munoz, S, Fuentes, N, Leonardo, D, Fuentealba, M, Herrera-Morande, A, Maturana, P, Villalobos, P, Garratt, R. | | Deposit date: | 2018-01-25 | | Release date: | 2018-02-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | ADP-Dependent Kinases From the Archaeal OrderMethanosarcinalesAdapt to Salt by a Non-canonical Evolutionarily Conserved Strategy.
Front Microbiol, 9, 2018
|
|
6MV6
 
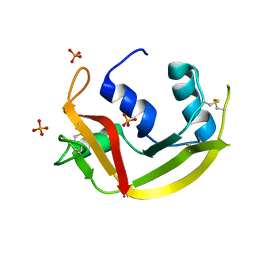 | | Crystal structure of RNAse 6 | | Descriptor: | PHOSPHATE ION, Ribonuclease K6 | | Authors: | Couture, J.-F, Doucet, N. | | Deposit date: | 2018-10-24 | | Release date: | 2019-11-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Insights into Structural and Dynamical Changes Experienced by Human RNase 6 upon Ligand Binding.
Biochemistry, 59, 2020
|
|
6MV7
 
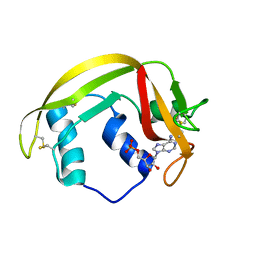 | | Crystal structure of RNAse 6 | | Descriptor: | ADENOSINE MONOPHOSPHATE, Ribonuclease K6 | | Authors: | Couture, J.-F, Doucet, N. | | Deposit date: | 2018-10-24 | | Release date: | 2019-11-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Insights into Structural and Dynamical Changes Experienced by Human RNase 6 upon Ligand Binding.
Biochemistry, 59, 2020
|
|
3A3A
 
 | |
6W62
 
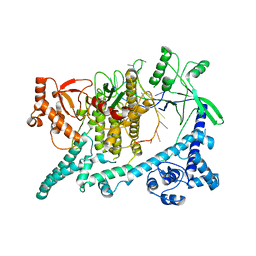 | | Cryo-EM structure of Cas12i-crRNA complex | | Descriptor: | Cas12i, crRNA | | Authors: | Chang, L, Li, Z, Zhang, H. | | Deposit date: | 2020-03-16 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Mechanisms for target recognition and cleavage by the Cas12i RNA-guided endonuclease.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7ACT
 
 | |
3R4F
 
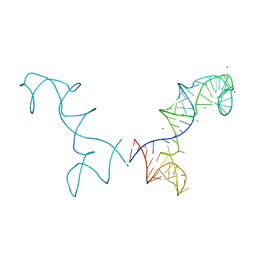 | | Prohead RNA | | Descriptor: | MAGNESIUM ION, pRNA | | Authors: | Ding, F, Lu, C, Zhano, W, Rajashankar, K.R, Anderson, D.L, Jardine, P.J, Grimes, S, Ke, A. | | Deposit date: | 2011-03-17 | | Release date: | 2011-04-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure and assembly of the essential RNA ring component of a viral DNA packaging motor.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3ZLA
 
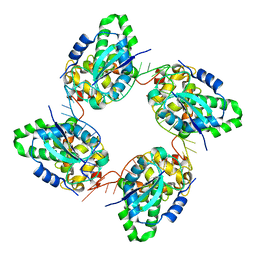 | | Crystal structure of the nucleocapsid protein from Bunyamwera virus bound to RNA | | Descriptor: | NUCLEOPROTEIN, RNA | | Authors: | Ariza, A, Tanner, S.J, Walter, C.T, Dent, K.C, Shepherd, D.A, Wu, W, Matthews, S.V, Hiscox, J.A, Green, T.J, Luo, M, Elliot, R.M, Ashcroft, A.E, Stonehouse, N.J, Ranson, N.A, Barr, J.N, Edwards, T.A. | | Deposit date: | 2013-01-29 | | Release date: | 2013-05-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Nucleocapsid Protein Structures from Orthobunyaviruses Reveal Insight Into Ribonucleoprotein Architecture and RNA Polymerization.
Nucleic Acids Res., 41, 2013
|
|
4V79
 
 | | E. coli 70S-fMetVal-tRNAVal-tRNAfMet complex in intermediate post-translocation state (post3b) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Blau, C, Bock, L.V, Schroder, G.F, Davydov, I, Fischer, N, Stark, H, Rodnina, M.V, Vaiana, A.C, Grubmuller, H. | | Deposit date: | 2013-10-14 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (15 Å) | | Cite: | Energy barriers and driving forces in tRNA translocation through the ribosome.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4V70
 
 | | E. coli 70S-fMetVal-tRNAVal-tRNAfMet complex in intermediate pre-translocation state (pre3) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Blau, C, Bock, L.V, Schroder, G.F, Davydov, I, Fischer, N, Stark, H, Rodnina, M.V, Vaiana, A.C, Grubmuller, H. | | Deposit date: | 2013-10-14 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (17 Å) | | Cite: | Energy barriers and driving forces in tRNA translocation through the ribosome.
Nat.Struct.Mol.Biol., 20, 2013
|
|
3JB7
 
 | | In situ structures of the segmented genome and RNA polymerase complex inside a dsRNA virus | | Descriptor: | CPV RNA-dependent RNA polymerase, CYTIDINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Zhang, X, Ding, K, Yu, X.K, Chang, W, Sun, J.C, Zhou, Z.H. | | Deposit date: | 2015-08-03 | | Release date: | 2015-10-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | In situ structures of the segmented genome and RNA polymerase complex inside a dsRNA virus.
Nature, 527, 2015
|
|
5JS2
 
 | | Human Argonaute-2 Bound to a Modified siRNA | | Descriptor: | MAGNESIUM ION, PHENOL, PHOSPHATE ION, ... | | Authors: | Schirle, N.T, MacRae, I.J. | | Deposit date: | 2016-05-07 | | Release date: | 2016-07-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.954 Å) | | Cite: | Structural Analysis of Human Argonaute-2 Bound to a Modified siRNA Guide.
J.Am.Chem.Soc., 138, 2016
|
|
5CYR
 
 | | Crystal structure of Thosea asigna virus RNA-dependent RNA polymerase (RdRP) complexed with ATP and ssRNA | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Ferrero, D.S, Buxaderas, M, Rodriguez, J.F, Verdaguer, N. | | Deposit date: | 2015-07-30 | | Release date: | 2015-11-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.504 Å) | | Cite: | The Structure of the RNA-Dependent RNA Polymerase of a Permutotetravirus Suggests a Link between Primer-Dependent and Primer-Independent Polymerases.
Plos Pathog., 11, 2015
|
|
5CX6
 
 | | Crystal structure of Thosea asigna virus RNA-dependent RNA polymerase (RdRP) complexed with CDP | | Descriptor: | CYTIDINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Ferrero, D.S, Buxaderas, M, Rodriguez, J.F, Verdaguer, N. | | Deposit date: | 2015-07-28 | | Release date: | 2015-11-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Structure of the RNA-Dependent RNA Polymerase of a Permutotetravirus Suggests a Link between Primer-Dependent and Primer-Independent Polymerases.
Plos Pathog., 11, 2015
|
|
8HSP
 
 | | E. coli 70S ribosome complexed with tRNA_Ile2 bearing L34 and t6A37 in classical state | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Akiyama, N, Ishiguro, K, Yokoyama, T, Shirouzu, M, Suzuki, T. | | Deposit date: | 2022-12-20 | | Release date: | 2024-04-03 | | Last modified: | 2025-02-12 | | Method: | ELECTRON MICROSCOPY (2.32 Å) | | Cite: | Structural insights into the decoding capability of isoleucine tRNAs with lysidine and agmatidine.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8HU1
 
 | | E. coli 70S ribosome complexed with tRNA_Ile2 bearing L34 and ct6A37 in classical state | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Akiyama, N, Ishiguro, K, Yokoyama, T, Shirouzu, M, Suzuki, T. | | Deposit date: | 2022-12-22 | | Release date: | 2024-04-03 | | Last modified: | 2025-02-12 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Structural insights into the decoding capability of isoleucine tRNAs with lysidine and agmatidine.
Nat.Struct.Mol.Biol., 31, 2024
|
|
