1WOX
 
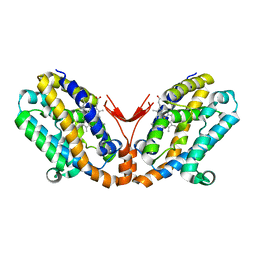 | | Crystal structure of heme oxygenase-2 from Synechocystis sp. PCC 6803 in complex with heme and NO | | Descriptor: | Heme oxygenase 2, NITRIC OXIDE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sugishima, M, Hagiwara, Y, Zhang, X, Yoshida, T, Migita, C.T, Fukuyama, K. | | Deposit date: | 2004-08-26 | | Release date: | 2005-03-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of dimeric heme oxygenase-2 from Synechocystis sp. PCC 6803 in complex with heme.
Biochemistry, 44, 2005
|
|
1WOY
 
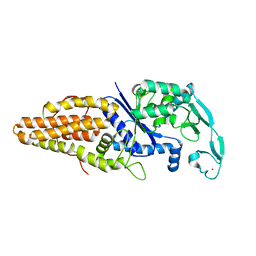 | |
1WOZ
 
 | | Crystal structure of uncharacterized protein ST1454 from Sulfolobus tokodaii | | Descriptor: | (20S)-2,5,8,11,14,17-HEXAMETHYL-3,6,9,12,15,18-HEXAOXAHENICOSANE-1,20-DIOL, 177aa long conserved hypothetical protein (ST1454) | | Authors: | Sasaki, T, Tanaka, Y, Yasutake, Y, Yao, M, Tanaka, I, Tsumoto, K, Kumagai, I. | | Deposit date: | 2004-08-27 | | Release date: | 2005-10-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structure of the uncharacterized protein ST1454 from Sulfolobus tokodaii.
To be Published
|
|
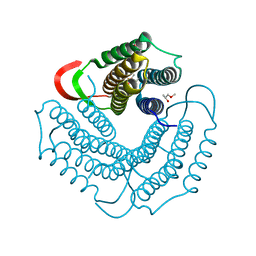
1WP0
 
 | | Human SCO1 | | Descriptor: | SCO1 protein homolog | | Authors: | Williams, J.C, Sue, C, Banting, G.S, Yang, H, Glerum, D.M, Hendrickson, W.A, Schon, E.A. | | Deposit date: | 2004-08-27 | | Release date: | 2005-01-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Human SCO1: IMPLICATIONS FOR REDOX SIGNALING BY A MITOCHONDRIAL CYTOCHROME c OXIDASE "ASSEMBLY" PROTEIN
J.Biol.Chem., 280, 2005
|
|
1WP1
 
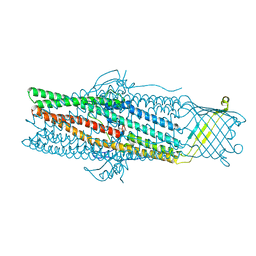 | | Crystal structure of the drug-discharge outer membrane protein, OprM | | Descriptor: | Outer membrane protein oprM | | Authors: | Akama, H, Kanemaki, M, Yoshimura, M, Tsukihara, T, Kashiwagi, T, Narita, S, Nakagawa, A, Nakae, T. | | Deposit date: | 2004-08-28 | | Release date: | 2004-11-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Crystal structure of the drug discharge outer membrane protein, OprM, of Pseudomonas aeruginosa: dual modes of membrane anchoring and occluded cavity end
J.Biol.Chem., 279, 2004
|
|
1WP4
 
 | |
1WP5
 
 | |
1WP6
 
 | | Crystal structure of maltohexaose-producing amylase from alkalophilic Bacillus sp.707. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, Glucan 1,4-alpha-maltohexaosidase, ... | | Authors: | Kanai, R, Haga, K, Akiba, T, Yamane, K, Harata, K. | | Deposit date: | 2004-08-31 | | Release date: | 2004-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Biochemical and crystallographic analyses of maltohexaose-producing amylase from alkalophilic Bacillus sp. 707
Biochemistry, 43, 2004
|
|
1WP7
 
 | | crystal structure of Nipah Virus fusion core | | Descriptor: | fusion protein | | Authors: | Xu, Y, Liu, Y, Lou, Z, Su, N, Bai, Z, Gao, G.F, Rao, Z. | | Deposit date: | 2004-08-31 | | Release date: | 2005-11-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Nipah Virus fusion core
To be Published
|
|
1WP8
 
 | | crystal structure of Hendra Virus fusion core | | Descriptor: | Fusion glycoprotein F0,Fusion glycoprotein F0 | | Authors: | Xu, Y, Liu, Y, Lou, Z, Su, N, Bai, Z, Gao, G.F, Rao, Z. | | Deposit date: | 2004-08-31 | | Release date: | 2005-11-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of Nipah and Hendra virus fusion core proteins
FEBS J., 273, 2006
|
|
1WP9
 
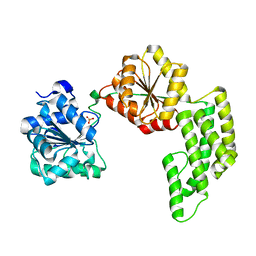 | | Crystal structure of Pyrococcus furiosus Hef helicase domain | | Descriptor: | ATP-dependent RNA helicase, putative, PHOSPHATE ION | | Authors: | Nishino, T, Komori, K, Tsuchiya, D, Ishino, Y, Morikawa, K. | | Deposit date: | 2004-08-31 | | Release date: | 2005-02-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure and Functional Implications of Pyrococcus furiosus Hef Helicase Domain Involved in Branched DNA Processing
Structure, 13, 2005
|
|
1WPA
 
 | |
1WPB
 
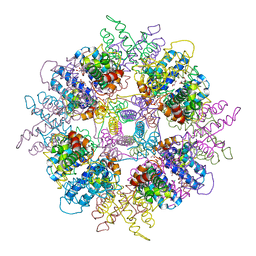 | | Structure of Escherichia coli yfbU gene product | | Descriptor: | CHLORIDE ION, GLYCEROL, hypothetical protein yfbU | | Authors: | Borek, D, Chen, Y, Zheng, M, Skarina, T, Savchenko, A, Edwards, A, Otwinowski, Z, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-09-01 | | Release date: | 2004-12-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Escherichia coli yfbU gene product
To be Published
|
|
1WPC
 
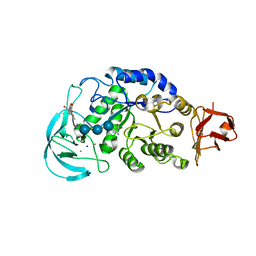 | | Crystal structure of maltohexaose-producing amylase complexed with pseudo-maltononaose | | Descriptor: | 4,6-dideoxy-alpha-D-xylo-hexopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, 4,6-dideoxy-alpha-D-xylo-hexopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-galactopyranose, 6-AMINO-4-HYDROXYMETHYL-CYCLOHEX-4-ENE-1,2,3-TRIOL, ... | | Authors: | Kanai, R, Haga, K, Akiba, T, Yamane, K, Harata, K. | | Deposit date: | 2004-09-01 | | Release date: | 2004-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Biochemical and crystallographic analyses of maltohexaose-producing amylase from alkalophilic Bacillus sp. 707
Biochemistry, 43, 2004
|
|
1WPD
 
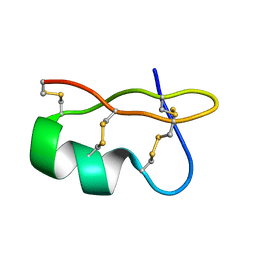 | | Evidence for domain-specific recognition of SK and Kv channels by MTX and HsTx1 scorpion toxins | | Descriptor: | Potassium channel toxin alpha-KTx 6.2,Potassium channel toxin alpha-KTx 6.3 | | Authors: | Regaya, I, Beeton, C, Ferrat, G, Andreotti, N, Chandy, G.K, Darbon, H, De Waard, M, Sabatier, J.M. | | Deposit date: | 2004-09-01 | | Release date: | 2004-10-19 | | Last modified: | 2019-12-18 | | Method: | SOLUTION NMR | | Cite: | Evidence for domain-specific recognition of SK and Kv channels by MTX and HsTx1 scorpion toxins
J.Biol.Chem., 279, 2004
|
|
1WPG
 
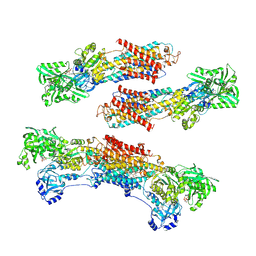 | | Crystal structure of the SR CA2+-ATPase with MGF4 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, OCTANOIC ACID [3S-[3ALPHA, ... | | Authors: | Toyoshima, C, Nomura, H, Tsuda, T. | | Deposit date: | 2004-09-02 | | Release date: | 2004-10-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Lumenal gating mechanism revealed in calcium pump crystal structures with phosphate analogues
Nature, 432, 2004
|
|
1WPI
 
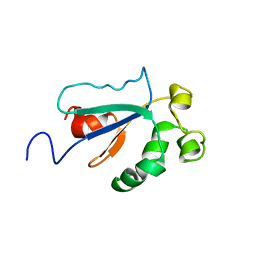 | | Solution NMR Structure of Protein YKR049C from Saccharomyces cerevisiae. Ontario Centre for Structural Proteomics target YST0250_1_133; Northeast Structural Genomics Consortium YTYst250 | | Descriptor: | Hypothetical 15.6 kDa protein in NAP1-TRK2 intergenic region | | Authors: | Jung, J.W, Yee, A, Arrowsmith, C.H, Lee, W, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-09-03 | | Release date: | 2005-09-13 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of YKR049C, a putative redox protein from Saccharomyces cerevisiae
J.Biochem.Mol.Biol., 38, 2005
|
|
1WPK
 
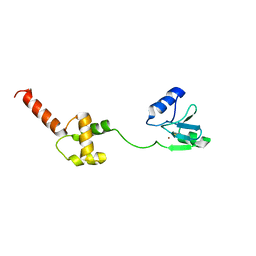 | | Methylated Form of N-terminal Transcriptional Regulator Domain of Escherichia Coli Ada Protein | | Descriptor: | ADA regulatory protein, ZINC ION | | Authors: | Takinowaki, H, Matsuda, Y, Yoshida, T, Kobayashi, Y, Ohkubo, T. | | Deposit date: | 2004-09-07 | | Release date: | 2005-09-13 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the methylated form of the N-terminal 16-kDa domain of Escherichia coli Ada protein
Protein Sci., 15, 2006
|
|
1WPL
 
 | | Crystal structure of the inhibitory form of rat GTP cyclohydrolase I/GFRP complex | | Descriptor: | 7,8-DIHYDROBIOPTERIN, GTP cyclohydrolase I, GTP cyclohydrolase I feedback regulatory protein, ... | | Authors: | Maita, N, Hatakeyama, K, Okada, K, Hakoshima, T. | | Deposit date: | 2004-09-08 | | Release date: | 2004-09-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of biopterin-induced inhibition of GTP cyclohydrolase I by GFRP, its feedback regulatory protein
J.Biol.Chem., 279, 2004
|
|
1WPM
 
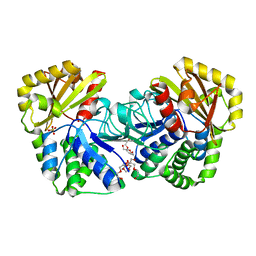 | | Structure of Bacillus subtilis inorganic pyrophosphatase | | Descriptor: | Manganese-dependent inorganic pyrophosphatase, SULFATE ION, TETRAETHYLENE GLYCOL | | Authors: | Fabrichniy, I.P, Lehtio, L, Salminen, A, Baykov, A.A, Lahti, R, Goldman, A. | | Deposit date: | 2004-09-09 | | Release date: | 2004-11-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Studies of Metal Ions in Family II Pyrophosphatases: The Requirement for a Janus Ion
Biochemistry, 43, 2004
|
|
1WPN
 
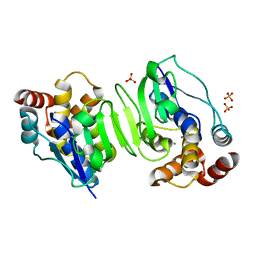 | | Crystal structure of the N-terminal core of Bacillus subtilis inorganic pyrophosphatase | | Descriptor: | MANGANESE (II) ION, Manganese-dependent inorganic pyrophosphatase, SULFATE ION | | Authors: | Fabrichniy, I.P, Lehtio, L, Salminen, A, Baykov, A.A, Lahti, R, Goldman, A. | | Deposit date: | 2004-09-09 | | Release date: | 2004-11-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural Studies of Metal Ions in Family II Pyrophosphatases: The Requirement for a Janus Ion
Biochemistry, 43, 2004
|
|
1WPO
 
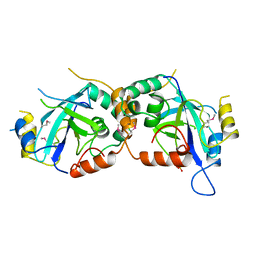 | | HYDROLYTIC ENZYME HUMAN CYTOMEGALOVIRUS PROTEASE | | Descriptor: | HUMAN CYTOMEGALOVIRUS PROTEASE, SULFATE ION | | Authors: | Tong, L. | | Deposit date: | 1996-07-23 | | Release date: | 1997-10-15 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A new serine-protease fold revealed by the crystal structure of human cytomegalovirus protease.
Nature, 383, 1996
|
|
1WPP
 
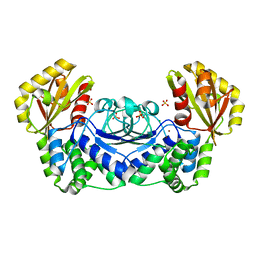 | | Structure of Streptococcus gordonii inorganic pyrophosphatase | | Descriptor: | CHLORIDE ION, Probable manganese-dependent inorganic pyrophosphatase, SULFATE ION, ... | | Authors: | Fabrichniy, I.P, Lehtio, L, Salminen, A, Baykov, A.A, Lahti, R, Goldman, A. | | Deposit date: | 2004-09-09 | | Release date: | 2004-11-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Studies of Metal Ions in Family II Pyrophosphatases: The Requirement for a Janus Ion
Biochemistry, 43, 2004
|
|
1WPQ
 
 | | Ternary Complex Of Glycerol 3-phosphate Dehydrogenase 1 with NAD and dihydroxyactone | | Descriptor: | 1,3-DIHYDROXYACETONEPHOSPHATE, Glycerol-3-phosphate dehydrogenase [NAD+], cytoplasmic, ... | | Authors: | Ou, X, Han, X, Rao, Z. | | Deposit date: | 2004-09-10 | | Release date: | 2006-04-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structures of Human Glycerol 3-phosphate Dehydrogenase 1 (GPD1)
J.Mol.Biol., 357, 2006
|
|
1WPR
 
 | | Crystal structure of RsbQ inhibited by PMSF | | Descriptor: | GLYCEROL, Sigma factor sigB regulation protein rsbQ, phenylmethanesulfonic acid | | Authors: | Kaneko, T, Tanaka, N, Kumasaka, T. | | Deposit date: | 2004-09-11 | | Release date: | 2005-02-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of RsbQ, a stress-response regulator in Bacillus subtilis
Protein Sci., 14, 2005
|
|
