1A54
 
 | | PHOSPHATE-BINDING PROTEIN MUTANT A197C LABELLED WITH A COUMARIN FLUOROPHORE AND BOUND TO DIHYDROGENPHOSPHATE ION | | Descriptor: | DIHYDROGENPHOSPHATE ION, N-[2-(1-MALEIMIDYL)ETHYL]-7-DIETHYLAMINOCOUMARIN-3-CARBOXAMIDE, Phosphate-binding protein PstS | | Authors: | Hirshberg, M, Henrick, K, Lloyd-Haire, L, Vasisht, N, Brune, M, Corrie, J.E.T, Webb, M.R. | | Deposit date: | 1998-02-19 | | Release date: | 1998-10-14 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of phosphate binding protein labeled with a coumarin fluorophore, a probe for inorganic phosphate.
Biochemistry, 37, 1998
|
|
7A6I
 
 | | Crystal Structure of EGFR-T790M/V948R in Complex with LDC8201 | | Descriptor: | Epidermal growth factor receptor, SULFATE ION, ~{N}-[5-[4-chloranyl-2-[4-(4-methylpiperazin-1-yl)phenyl]-1~{H}-pyrrolo[2,3-b]pyridin-3-yl]-2-methyl-phenyl]propanamide | | Authors: | Niggenaber, J, Mueller, M.P, Rauh, D. | | Deposit date: | 2020-08-25 | | Release date: | 2022-02-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Insight into Targeting Exon20 Insertion Mutations of the Epidermal Growth Factor Receptor with Wild Type-Sparing Inhibitors.
J.Med.Chem., 65, 2022
|
|
7SIH
 
 | | Crystal Structure of HLA B*3503 in complex with NPDIVIYQY, an 9-mer epitope from HIV-I | | Descriptor: | Beta-2-microglobulin, GLYCEROL, MHC class I antigen, ... | | Authors: | Gras, S, Lobos, C.A, Chatzileontiadou, D.S.M. | | Deposit date: | 2021-10-14 | | Release date: | 2022-11-23 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular insights into the HLA-B35 molecules' classification associated with HIV control.
Immunol.Cell.Biol., 102, 2024
|
|
1A7E
 
 | |
4WMC
 
 | | OXA-48 covalent complex with Avibactam inhibitor | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase, CARBON DIOXIDE | | Authors: | Mangani, S, Benvenuti, M, Docquier, J.D. | | Deposit date: | 2014-10-08 | | Release date: | 2014-12-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular Basis of Selective Inhibition and Slow Reversibility of Avibactam against Class D Carbapenemases: A Structure-Guided Study of OXA-24 and OXA-48.
Acs Chem.Biol., 10, 2015
|
|
7A6J
 
 | | Crystal Structure of EGFR-T790M/V948R in Complex with Poziotinib | | Descriptor: | 1,2-ETHANEDIOL, 1-[4-[4-[[3,4-bis(chloranyl)-2-fluoranyl-phenyl]amino]-7-methoxy-quinazolin-6-yl]oxypiperidin-1-yl]propan-1-one, Epidermal growth factor receptor, ... | | Authors: | Niggenaber, J, Mueller, M.P, Rauh, D. | | Deposit date: | 2020-08-25 | | Release date: | 2022-02-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insight into Targeting Exon20 Insertion Mutations of the Epidermal Growth Factor Receptor with Wild Type-Sparing Inhibitors.
J.Med.Chem., 65, 2022
|
|
1A57
 
 | | THE THREE-DIMENSIONAL STRUCTURE OF A HELIX-LESS VARIANT OF INTESTINAL FATTY ACID BINDING PROTEIN, NMR, 20 STRUCTURES | | Descriptor: | INTESTINAL FATTY ACID-BINDING PROTEIN | | Authors: | Steele, R.A, Emmert, D.A, Kao, J, Hodsdon, M.E, Frieden, C, Cistola, D.P. | | Deposit date: | 1998-02-20 | | Release date: | 1998-05-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional structure of a helix-less variant of intestinal fatty acid-binding protein.
Protein Sci., 7, 1998
|
|
1ACD
 
 | | V32D/F57H MUTANT OF MURINE ADIPOCYTE LIPID BINDING PROTEIN | | Descriptor: | ADIPOCYTE LIPID BINDING PROTEIN | | Authors: | Ory, J, Kane, C.D, Simpson, M, Banaszak, L.J, Bernlohr, D.A. | | Deposit date: | 1997-02-06 | | Release date: | 1997-06-16 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Biochemical and crystallographic analyses of a portal mutant of the adipocyte lipid-binding protein.
J.Biol.Chem., 272, 1997
|
|
1A55
 
 | | PHOSPHATE-BINDING PROTEIN MUTANT A197C | | Descriptor: | DIHYDROGENPHOSPHATE ION, PHOSPHATE-BINDING PROTEIN | | Authors: | Hirshberg, M, Henrick, K, Lloyd-Haire, L, Vasisht, N, Brune, M, Corrie, J.E.T, Webb, M.R. | | Deposit date: | 1998-02-19 | | Release date: | 1998-10-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of phosphate binding protein labeled with a coumarin fluorophore, a probe for inorganic phosphate.
Biochemistry, 37, 1998
|
|
7A6K
 
 | | Crystal Structure of EGFR-T790M/V948R in Complex with TAK-788 | | Descriptor: | Epidermal growth factor receptor, SULFATE ION, propan-2-yl 2-[[4-[2-(dimethylamino)ethyl-methyl-amino]-2-methoxy-5-(propanoylamino)phenyl]amino]-4-(1-methylindol-3-yl)pyrimidine-5-carboxylate | | Authors: | Niggenaber, J, Mueller, M.P, Rauh, D. | | Deposit date: | 2020-08-25 | | Release date: | 2022-02-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insight into Targeting Exon20 Insertion Mutations of the Epidermal Growth Factor Receptor with Wild Type-Sparing Inhibitors.
J.Med.Chem., 65, 2022
|
|
7SIG
 
 | | Crystal Structure of HLA B*3501 in complex with NPDIVIYQY, an 9-mer epitope from HIV-I | | Descriptor: | Beta-2-microglobulin, CITRATE ANION, MHC class I antigen, ... | | Authors: | Gras, S, Lobos, C.A, Chatzileontiadou, D.S.M. | | Deposit date: | 2021-10-14 | | Release date: | 2022-11-23 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.741 Å) | | Cite: | Molecular insights into the HLA-B35 molecules' classification associated with HIV control.
Immunol.Cell.Biol., 102, 2024
|
|
1A8M
 
 | | TUMOR NECROSIS FACTOR ALPHA, R31D MUTANT | | Descriptor: | TUMOR NECROSIS FACTOR ALPHA | | Authors: | Reed, C, Fu, Z.-Q, Wu, J, Xue, Y.-N, Harrison, R.W, Chen, M.-J, Weber, I.T. | | Deposit date: | 1998-03-27 | | Release date: | 1998-06-17 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of TNF-alpha mutant R31D with greater affinity for receptor R1 compared with R2.
Protein Eng., 10, 1997
|
|
7SIF
 
 | | Crystal Structure of HLA B*3505 in complex with NPDIVIYQY, an 9-mer epitope from HIV-I | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-2-microglobulin, ... | | Authors: | Gras, S, Lobos, C.A, Chatzileontiadou, D.S.M. | | Deposit date: | 2021-10-14 | | Release date: | 2022-11-23 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Molecular insights into the HLA-B35 molecules' classification associated with HIV control.
Immunol.Cell.Biol., 102, 2024
|
|
12AS
 
 | | ASPARAGINE SYNTHETASE MUTANT C51A, C315A COMPLEXED WITH L-ASPARAGINE AND AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, ASPARAGINE, ASPARAGINE SYNTHETASE | | Authors: | Nakatsu, T, Kato, H, Oda, J. | | Deposit date: | 1997-12-02 | | Release date: | 1998-12-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of asparagine synthetase reveals a close evolutionary relationship to class II aminoacyl-tRNA synthetase.
Nat.Struct.Biol., 5, 1998
|
|
6Y0H
 
 | | High resolution structure of GH11 xylanase from Nectria haematococca | | Descriptor: | Endo-1,4-beta-xylanase | | Authors: | Andaleeb, H, Betzel, C, Perbandt, M, Brognaro, H. | | Deposit date: | 2020-02-07 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | High-resolution crystal structure and biochemical characterization of a GH11 endoxylanase from Nectria haematococca.
Sci Rep, 10, 2020
|
|
1A6L
 
 | | T14C MUTANT OF AZOTOBACTER VINELANDII FDI | | Descriptor: | FE3-S4 CLUSTER, FERREDOXIN, IRON/SULFUR CLUSTER | | Authors: | Gao-Sheridan, H.S, Kemper, M.A, Khayat, R, Armstrong, F.A, Prasad, G.S, Sridhar, V, Stout, C.D, Burgess, B.K. | | Deposit date: | 1998-02-26 | | Release date: | 1998-05-27 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A T14C variant of Azotobacter vinelandii ferredoxin I undergoes facile [3Fe-4S]0 to [4Fe-4S]2+ conversion in vitro but not in vivo.
J.Biol.Chem., 273, 1998
|
|
1A8K
 
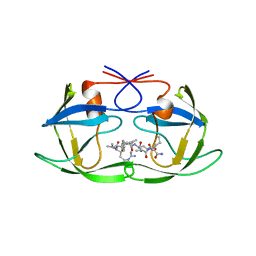 | | CRYSTALLOGRAPHIC ANALYSIS OF HUMAN IMMUNODEFICIENCY VIRUS 1 PROTEASE WITH AN ANALOG OF THE CONSERVED CA-P2 SUBSTRATE: INTERACTIONS WITH FREQUENTLY OCCURRING GLUTAMIC ACID RESIDUE AT P2' POSITION OF SUBSTRATES | | Descriptor: | HIV PROTEASE, N-[(2R)-2-({N~5~-[amino(iminio)methyl]-L-ornithyl-L-valyl}amino)-4-methylpentyl]-L-phenylalanyl-L-alpha-glutamyl-L-alanyl-L-norleucinamide | | Authors: | Weber, I.T, Wu, J, Adomat, J, Harrison, R.W, Kimmel, A.R, Wondrak, E.M, Louis, J.M. | | Deposit date: | 1998-03-27 | | Release date: | 1999-01-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic analysis of human immunodeficiency virus 1 protease with an analog of the conserved CA-p2 substrate -- interactions with frequently occurring glutamic acid residue at P2' position of substrates.
Eur.J.Biochem., 249, 1997
|
|
5KO6
 
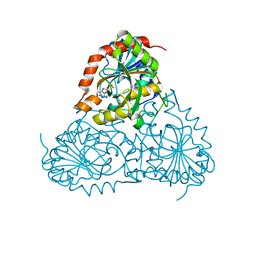 | | Crystal Structure of Isoform 2 of Purine Nucleoside Phosphorylase from Schistosoma mansoni in complex with cytosine and ribose-1-phosphate | | Descriptor: | 1-O-phosphono-alpha-D-ribofuranose, 6-AMINOPYRIMIDIN-2(1H)-ONE, Purine nucleoside phosphorylase | | Authors: | Torini, J.R, Romanello, L, Bird, L, Owens, R, Brandao-Neto, J, Pereira, H.M. | | Deposit date: | 2016-06-29 | | Release date: | 2017-08-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | The molecular structure of Schistosoma mansoni PNP isoform 2 provides insights into the nucleoside selectivity of PNPs.
PLoS ONE, 13, 2018
|
|
3LU8
 
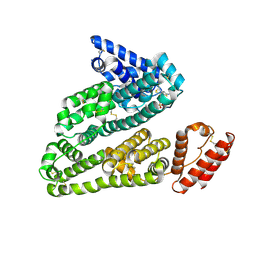 | | Human serum albumin in complex with compound 3 | | Descriptor: | N-[5-(5-{[(2,4-dimethyl-1,3-thiazol-5-yl)sulfonyl]amino}-6-fluoropyridin-3-yl)-4-methyl-1,3-thiazol-2-yl]acetamide, Serum albumin | | Authors: | Buttar, D, Colclough, N, Gerhardt, S, MacFaul, P.A, Phillips, S.D, Plowright, A, Whittamore, P, Tam, K, Maskos, K, Steinbacher, S, Steuber, H. | | Deposit date: | 2010-02-17 | | Release date: | 2010-10-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A combined spectroscopic and crystallographic approach to probing drug-human serum albumin interactions
Bioorg.Med.Chem., 18, 2010
|
|
1AAX
 
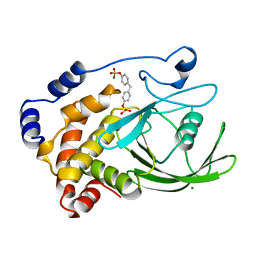 | | CRYSTAL STRUCTURE OF PROTEIN TYROSINE PHOSPHATASE 1B COMPLEXED WITH TWO BIS(PARA-PHOSPHOPHENYL)METHANE (BPPM) MOLECULES | | Descriptor: | 4-PHOSPHONOOXY-PHENYL-METHYL-[4-PHOSPHONOOXY]BENZEN, MAGNESIUM ION, PROTEIN TYROSINE PHOSPHATASE 1B | | Authors: | Puius, Y.A, Zhao, Y, Sullivan, M, Lawrence, D, Almo, S.C, Zhang, Z.-Y. | | Deposit date: | 1997-01-16 | | Release date: | 1998-03-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of a second aryl phosphate-binding site in protein-tyrosine phosphatase 1B: a paradigm for inhibitor design.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
1A0G
 
 | | L201A MUTANT OF D-AMINO ACID AMINOTRANSFERASE COMPLEXED WITH PYRIDOXAMINE-5'-PHOSPHATE | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, D-AMINO ACID AMINOTRANSFERASE | | Authors: | Sugio, S, Kashima, A, Kishimoto, K, Peisach, D, Petsko, G.A, Ringe, D, Yoshimura, T, Esaki, N. | | Deposit date: | 1997-11-30 | | Release date: | 1998-06-03 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of L201A mutant of D-amino acid aminotransferase at 2.0 A resolution: implication of the structural role of Leu201 in transamination.
Protein Eng., 11, 1998
|
|
1A6I
 
 | | TET REPRESSOR, CLASS D VARIANT | | Descriptor: | TETRACYCLINE REPRESSOR PROTEIN CLASS D | | Authors: | Orth, P, Cordes, F, Schnappinger, D, Hillen, W, Saenger, W, Hinrichs, W. | | Deposit date: | 1998-02-25 | | Release date: | 1999-03-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Conformational changes of the Tet repressor induced by tetracycline trapping.
J.Mol.Biol., 279, 1998
|
|
1A9M
 
 | | G48H MUTANT OF HIV-1 PROTEASE IN COMPLEX WITH A PEPTIDIC INHIBITOR U-89360E | | Descriptor: | HIV-1 PROTEASE, N-[[1-[N-ACETAMIDYL]-[1-CYCLOHEXYLMETHYL-2-HYDROXY-4-ISOPROPYL]-BUT-4-YL]-CARBONYL]-GLUTAMINYL-ARGINYL-AMIDE | | Authors: | Hong, L, Zhang, X.-J, Foundling, S, Hartsuck, J.A, Tang, J. | | Deposit date: | 1998-04-08 | | Release date: | 1998-06-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of a G48H mutant of HIV-1 protease explains how glycine-48 replacements produce mutants resistant to inhibitor drugs.
FEBS Lett., 420, 1997
|
|
1A9I
 
 | |
103D
 
 | |
