6VH6
 
 | |
2UZC
 
 | | Structure of human PDLIM5 in complex with the C-terminal peptide of human alpha-actinin-1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, PDZ AND LIM DOMAIN 5 | | Authors: | Bunkoczi, G, Elkins, J, Salah, E, Burgess-Brown, N, Papagrigoriou, E, Pike, A.C.W, Turnbull, A, Gileadi, O, von Delft, F, Arrowsmith, C.H, Edwards, A, Sundstrom, M, Weigelt, J, Doyle, D. | | Deposit date: | 2007-04-27 | | Release date: | 2007-05-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Unusual binding interactions in PDZ domain crystal structures help explain binding mechanisms.
Protein Sci., 19, 2010
|
|
5BUW
 
 | |
6RSB
 
 | |
5AP0
 
 | | Naturally Occurring Mutations in the MPS1 Gene Predispose Cells to Kinase Inhibitor Drug Resistance. | | Descriptor: | 1,2-ETHANEDIOL, 9-CYCLOPENTYL-2-[[2-METHOXY-4-[(1-METHYLPIPERIDIN-4-YL)OXY]-PHENYL]AMINO]-7-METHYL-7,9-DIHYDRO-8H-PURIN-8-ONE, DIMETHYL SULFOXIDE, ... | | Authors: | Gurden, M.D, Westwood, I.M, Faisal, A, Naud, S, Cheung, K.J, McAndrew, C, Wood, A, Schmitt, J, Boxall, K, Mak, G, Workman, P, Burke, R, Hoelder, S, Blagg, J, van Montfort, R.L.M, Linardopoulos, S. | | Deposit date: | 2015-09-14 | | Release date: | 2015-09-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Naturally Occurring Mutations in the Mps1 Gene Predispose Cells to Kinase Inhibitor Drug Resistance.
Cancer Res., 75, 2015
|
|
2VBG
 
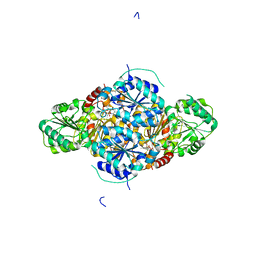 | | The complex structure of the branched-chain keto acid decarboxylase (KdcA) from Lactococcus lactis with 2R-1-hydroxyethyl-deazaThDP | | Descriptor: | 2-{4-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-5-[(1R)-1-HYDROXYETHYL]-3-METHYL-2-THIENYL}ETHYL TRIHYDROGEN DIPHOSPHATE, BRANCHED-CHAIN ALPHA-KETOACID DECARBOXYLASE, MAGNESIUM ION | | Authors: | Berthold, C.L, Gocke, D, Wood, M.D, Leeper, F, Pohl, M, Schneider, G. | | Deposit date: | 2007-09-12 | | Release date: | 2007-11-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the Branched-Chain Keto Acid Decarboxylase (Kdca) from Lactococcus Lactis Provides Insights Into the Structural Basis for the Chemo- and Enantioselective Carboligation Reaction
Acta Crystallogr.,Sect.D, 63, 2007
|
|
5EFD
 
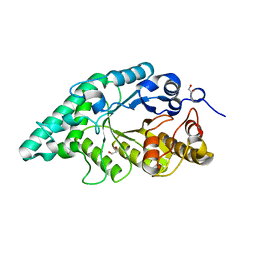 | | Crystal structure of a surface pocket creating mutant (W6A) of an alkali thermostable GH10 xylanase from Bacillus sp. NG-27 | | Descriptor: | 1,2-ETHANEDIOL, Beta-xylanase, CHLORIDE ION, ... | | Authors: | Mahanta, P, Bhardwaj, A, Reddy, V.S, Ramakumar, S. | | Deposit date: | 2015-10-23 | | Release date: | 2016-10-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.674 Å) | | Cite: | Small Glycols Discover Cryptic Pockets on Proteins for Fragment-Based Approaches.
J.Chem.Inf.Model., 2021
|
|
3ZH7
 
 | | The structure of crystal form II of Haemophilus influenzae protein E | | Descriptor: | 1,2-ETHANEDIOL, PROTEIN E | | Authors: | Singh, B, Al-Jubair, T, Riesbeck, K, Thunnissen, M.M.G.M. | | Deposit date: | 2012-12-20 | | Release date: | 2013-04-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | The Unique Structure of Haemophilus Influenzae Protein E Reveals Multiple Binding Sites for Host Factors.
Infect.Immun., 81, 2013
|
|
6AD5
 
 | |
7L3P
 
 | |
7VX2
 
 | | Crystal Structure of the Y53F/N55A/I80F/L114V/I116V mutant of LEH | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Qu, G, Li, X, Sun, Z.T, Han, X, Liu, W.D. | | Deposit date: | 2021-11-12 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.485 Å) | | Cite: | Rational enzyme design for enabling biocatalytic Baldwin cyclization and asymmetric synthesis of chiral heterocycles.
Nat Commun, 13, 2022
|
|
7VWD
 
 | | Crystal Structure of the Y53F/N55A mutant of LEH | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Qu, G, Li, X, Sun, Z.T, Han, X, Liu, W.D. | | Deposit date: | 2021-11-10 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.153 Å) | | Cite: | Rational enzyme design for enabling biocatalytic Baldwin cyclization and asymmetric synthesis of chiral heterocycles.
Nat Commun, 13, 2022
|
|
6S88
 
 | | Fumarate hydratase of Mycobacterium tuberculosis in complex with formate and allosteric modulator N-(2-Methoxy-5-((1,2,4,5-tetrahydro-3H-benzo[d]azepin-3-yl)sulfonyl)phenyl)-2-(4-oxo-3,4-dihydrophthalazin-1-yl)acetamide | | Descriptor: | FORMIC ACID, Fumarate hydratase class II, MAGNESIUM ION, ... | | Authors: | Whitehouse, A.J, Libardo, M.D, Kasbekar, M, Brear, P, Fischer, G, Thomas, C.J, Barry, C.E, Boshoff, H.I, Coyne, A.G, Abell, C. | | Deposit date: | 2019-07-08 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Targeting of Fumarate Hydratase fromMycobacterium tuberculosisUsing Allosteric Inhibitors with a Dimeric-Binding Mode.
J.Med.Chem., 62, 2019
|
|
2V8C
 
 | | Mouse Profilin IIa in complex with the proline-rich domain of VASP | | Descriptor: | GLYCEROL, ISOPROPYL ALCOHOL, PROFILIN-2, ... | | Authors: | Kursula, P, Downer, J, Witke, W, Wilmanns, M. | | Deposit date: | 2007-08-06 | | Release date: | 2007-12-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | High-Resolution Structural Analysis of Mammalian Profilin 2A Complex Formation with Two Physiological Ligands: The Formin Homology 1 Domain of Mdia1 and the Proline-Rich Domain of Vasp.
J.Mol.Biol., 375, 2008
|
|
3M81
 
 | |
3C44
 
 | | Crystal structure of HIV-1 subtype F DIS extended duplex RNA bound to paromomycin | | Descriptor: | CHLORIDE ION, HIV-1 subtype F genomic RNA, PAROMOMYCIN, ... | | Authors: | Freisz, S, Ennifar, E, Dumas, P. | | Deposit date: | 2008-01-29 | | Release date: | 2008-05-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Binding of aminoglycoside antibiotics to the duplex form of the HIV-1 genomic RNA dimerization initiation site.
Angew.Chem.Int.Ed.Engl., 47, 2008
|
|
5HZD
 
 | | RNA Editing TUTase 1 from Trypanosoma brucei | | Descriptor: | 3' terminal uridylyl transferase, CHLORIDE ION, SULFATE ION, ... | | Authors: | Thore, S, Rajappa, L.T. | | Deposit date: | 2016-02-02 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | RNA Editing TUTase 1: structural foundation of substrate recognition, complex interactions and drug targeting.
Nucleic Acids Res., 44, 2016
|
|
2SN3
 
 | | STRUCTURE OF SCORPION TOXIN VARIANT-3 AT 1.2 ANGSTROMS RESOLUTION | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, SCORPION NEUROTOXIN (VARIANT 3) | | Authors: | Zhao, B, Carson, M, Ealick, S.E, Bugg, C.E. | | Deposit date: | 1992-02-20 | | Release date: | 1994-01-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of scorpion toxin variant-3 at 1.2 A resolution.
J.Mol.Biol., 227, 1992
|
|
2RIT
 
 | | Unliganded B-specific-1,3-galactosyltransferase (GTB) | | Descriptor: | GLYCEROL, Glycoprotein-fucosylgalactoside alpha-galactosyltransferase | | Authors: | Evans, S.V, Alfaro, J.A. | | Deposit date: | 2007-10-12 | | Release date: | 2008-02-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | ABO(H) blood group A and B glycosyltransferases recognize substrate via specific conformational changes.
J.Biol.Chem., 283, 2008
|
|
2RJ7
 
 | |
6RUJ
 
 | |
7D6B
 
 | | Crystal structure of Oryza sativa Os4BGlu18 monolignol beta-glucosidase with delta-gluconolactone | | Descriptor: | Beta-glucosidase 18, D-glucono-1,5-lactone, GLYCEROL, ... | | Authors: | Baiya, S, Pengthaisong, S, Ketudat Cairns, J.R. | | Deposit date: | 2020-09-29 | | Release date: | 2021-01-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural analysis of rice Os4BGlu18 monolignol beta-glucosidase.
Plos One, 16, 2021
|
|
6FTW
 
 | | Crystal structure of human phosphodiesterase 4D2 catalytic domain with inhibitor NPD-048 | | Descriptor: | 1,2-ETHANEDIOL, 3-{5-[(4aR,8aS)-3-cycloheptyl-4-oxo-3,4,4a,5,8,8a-hexahydrophthalazin-1-yl]-2-methoxyphenyl}prop-2-ynamide, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Singh, A.K, Brown, D.G. | | Deposit date: | 2018-02-24 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Alkynamide phthalazinones as a new class of TbrPDEB1 inhibitors.
Bioorg.Med.Chem., 27, 2019
|
|
7WSV
 
 | | Cryo-EM structure of the N-terminal deletion mutant of human pannexin-1 in a nanodisc | | Descriptor: | Pannexin-1 | | Authors: | Kuzuya, M, Hirano, H, Hayashida, K, Watanabe, M, Kobayashi, K, Tani, K, Fujiyoshi, Y, Oshima, A. | | Deposit date: | 2022-02-01 | | Release date: | 2022-02-16 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structures of human pannexin-1 in nanodiscs reveal gating mediated by dynamic movement of the N terminus and phospholipids.
Sci.Signal., 15, 2022
|
|
2RJ8
 
 | |
