6OKG
 
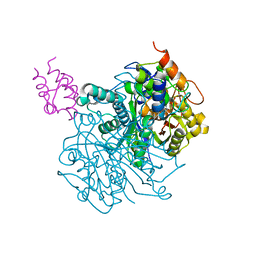 | | Crosslinked Crystal Structure of Type II Fatty Acid Synthase Ketosynthase, FabF, and C16-crypto Acyl Carrier Protein, AcpP | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 2, Acyl carrier protein, N-[2-(hexadecanoylamino)ethyl]-N~3~-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alaninamide, ... | | Authors: | Mindrebo, J.T, Kim, W.E, Bartholow, T.G, Chen, A, Davis, T.D, La Clair, J, Burkart, M.D, Noel, J.P. | | Deposit date: | 2019-04-12 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Gating mechanism of elongating beta-ketoacyl-ACP synthases.
Nat Commun, 11, 2020
|
|
6KKL
 
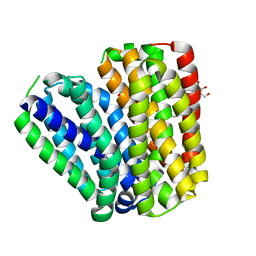 | | Crystal structure of Drug:Proton Antiporter-1 (DHA1) Family SotB, in the inward conformation (H115N mutant) | | Descriptor: | Sugar efflux transporter, nonyl beta-D-glucopyranoside | | Authors: | Xiao, Q.J, Sun, B, Zuo, Y.X, Guo, L, He, J.H, Deng, D. | | Deposit date: | 2019-07-26 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.654 Å) | | Cite: | Visualizing the nonlinear changes of a drug-proton antiporter from inward-open to occluded state.
Biochem.Biophys.Res.Commun., 534, 2021
|
|
6FIN
 
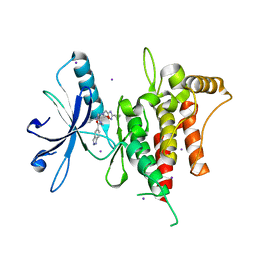 | | DDR1, 3-[(3-cyclopropyl-1,2,4-oxadiazol-5-yl)methyl]-8-(1H-indazole-5-carbonyl)-1-phenyl-1,3,8-triazaspiro[4.5]decan-4-one, 1.670A, P1211, Rfree=22.8% | | Descriptor: | 3-[(3-cyclopropyl-1,2,4-oxadiazol-5-yl)methyl]-8-(1~{H}-indazol-5-ylcarbonyl)-1-phenyl-1,3,8-triazaspiro[4.5]decan-4-one, Epithelial discoidin domain-containing receptor 1, IODIDE ION | | Authors: | Stihle, M, Richter, H, Benz, J, Kuhn, B, Rudolph, M.G. | | Deposit date: | 2018-01-19 | | Release date: | 2018-11-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | DNA-Encoded Library-Derived DDR1 Inhibitor Prevents Fibrosis and Renal Function Loss in a Genetic Mouse Model of Alport Syndrome.
Acs Chem.Biol., 14, 2019
|
|
5H7W
 
 | |
6FIQ
 
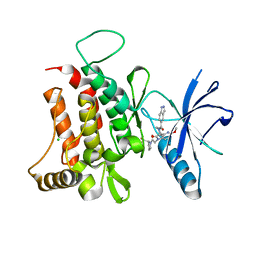 | | DDR1, 1-(1H-indazole-5-carbonyl)-5'-methoxy-1'-[2-oxo-2-[(2S)-2-(trifluoromethyl)pyrrolidin-1-yl]ethyl]spiro[piperidine-4,3'-pyrrolo[3,2-b]pyridine]-2'-one, 1.790A, P212121, Rfree=23.8% | | Descriptor: | 1-(1~{H}-indazol-5-ylcarbonyl)-5'-methoxy-1'-[2-oxidanylidene-2-[(2~{S})-2-(trifluoromethyl)pyrrolidin-1-yl]ethyl]spiro[piperidine-4,3'-pyrrolo[3,2-b]pyridine]-2'-one, CHLORIDE ION, Epithelial discoidin domain-containing receptor 1 | | Authors: | Stihle, M, Richter, H, Benz, J, Kuhn, B, Rudolph, M.G. | | Deposit date: | 2018-01-19 | | Release date: | 2018-11-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | DNA-Encoded Library-Derived DDR1 Inhibitor Prevents Fibrosis and Renal Function Loss in a Genetic Mouse Model of Alport Syndrome.
Acs Chem.Biol., 14, 2019
|
|
6OKP
 
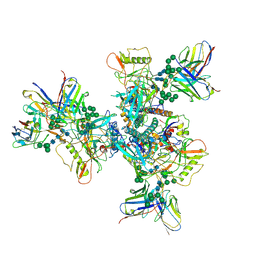 | | B41 SOSIP.664 in complex with the silent-face antibody SF12 and V3-targeting antibody 10-1074 | | Descriptor: | 10-1074 Heavy Chain,10-1074 Heavy Chain, 10-1074 Light Chain,10-1074 Light Chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Barnes, C.O, Bjorkman, P.J. | | Deposit date: | 2019-04-14 | | Release date: | 2019-06-05 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Broad and Potent Neutralizing Antibodies Recognize the Silent Face of the HIV Envelope.
Immunity, 50, 2019
|
|
5K6Z
 
 | | Sidekick chimera containing sidekick-2 immunoglobulin domains 1-2 and sidekick-1 immunoglobulin domains 3-4 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Goodman, K.M, Mannepalli, S, Honig, B, Shapiro, L. | | Deposit date: | 2016-05-25 | | Release date: | 2016-09-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular basis of sidekick-mediated cell-cell adhesion and specificity.
Elife, 5, 2016
|
|
6OLT
 
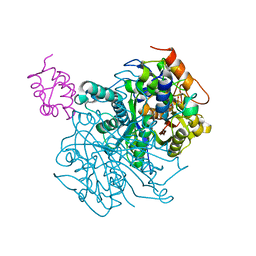 | | Crosslinked Crystal Structure of Type II Fatty Acid Synthase Ketosynthase, FabF, and C12-crypto Acyl Carrier Protein, AcpP | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 2, Acyl carrier protein, N-[2-(dodecanoylamino)ethyl]-N~3~-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alaninamide | | Authors: | Mindrebo, J.T, Kim, W.E, Bartholow, T.G, Chen, A, Davis, T.D, La Clair, J, Burkart, M.D, Noel, J.P. | | Deposit date: | 2019-04-17 | | Release date: | 2020-04-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Gating mechanism of elongating beta-ketoacyl-ACP synthases.
Nat Commun, 11, 2020
|
|
8CRR
 
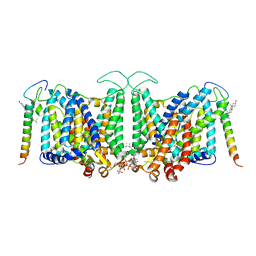 | | Local refinement of Band 3-III transmembrane domains, class 1 of erythrocyte ankyrin-1 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Band 3 anion transport protein, CHOLESTEROL, ... | | Authors: | Vallese, F, Kim, K, Yen, L.Y, Johnston, J.D, Noble, A.J, Cali, T, Clarke, O.B. | | Deposit date: | 2022-05-11 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Architecture of the human erythrocyte ankyrin-1 complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
8DCD
 
 | | RNA ligase RtcB from Pyrococcus horikoshii in complex with Zn2+ and GTP | | Descriptor: | CHLORIDE ION, GLYCEROL, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Jacewicz, A, Dantuluri, S, Shuman, S. | | Deposit date: | 2022-06-16 | | Release date: | 2022-10-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structures of RNA ligase RtcB in complexes with divalent cations and GTP.
Rna, 28, 2022
|
|
6FEW
 
 | | DDR1, 2-[8-(1H-indazole-5-carbonyl)-4-oxo-1-phenyl-1,3,8-triazaspiro[4.5]decan-3-yl]-N-methylacetamide, 1.440A, P1211, Rfree=24.1% | | Descriptor: | 2-[8-(2~{H}-indazol-5-ylcarbonyl)-4-oxidanylidene-1-phenyl-1,3,8-triazaspiro[4.5]decan-3-yl]-~{N}-methyl-ethanamide, Epithelial discoidin domain-containing receptor 1 | | Authors: | Stihle, M, Richter, H, Benz, J, Kuhn, B, Rudolph, M.G. | | Deposit date: | 2018-01-03 | | Release date: | 2018-11-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | DNA-Encoded Library-Derived DDR1 Inhibitor Prevents Fibrosis and Renal Function Loss in a Genetic Mouse Model of Alport Syndrome.
Acs Chem.Biol., 14, 2019
|
|
6FFI
 
 | | Crystal Structure of mGluR5 in complex with MMPEP at 2.2 A | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-[2-(3-methoxyphenyl)ethynyl]-6-methyl-pyridine, Metabotropic glutamate receptor 5,Endolysin,Metabotropic glutamate receptor 5, ... | | Authors: | Christopher, J.A, Orgovan, Z, Congreve, M, Dore, A.S, Errey, J.C, Marshall, F.H, Mason, J.S, Okrasa, K, Rucktooa, P, Serrano-Vega, M.J, Ferenczy, G.G, Keseru, G.M. | | Deposit date: | 2018-01-08 | | Release date: | 2018-03-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Based Optimization Strategies for G Protein-Coupled Receptor (GPCR) Allosteric Modulators: A Case Study from Analyses of New Metabotropic Glutamate Receptor 5 (mGlu5) X-ray Structures.
J.Med.Chem., 62, 2019
|
|
8CRT
 
 | | Local refinement of Rh trimer, glycophorin B and Band3-III transmembrane region, class 1a of erythrocyte ankyrin-1 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ammonium transporter Rh type A, Band 3 anion transport protein, ... | | Authors: | Vallese, F, Kim, K, Yen, L.Y, Johnston, J.D, Noble, A.J, Cali, T, Clarke, O.B. | | Deposit date: | 2022-05-11 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Architecture of the human erythrocyte ankyrin-1 complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
5JPG
 
 | | Rat Galectin 5 with lactose | | Descriptor: | Galectin-5, SODIUM ION, beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Romero, A, Ruiz, F.M. | | Deposit date: | 2016-05-03 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Characterization of Rat Galectin-5, an N-Tailed Monomeric Proto-Type-like Galectin.
Biomolecules, 11, 2021
|
|
5VTA
 
 | | Co-Crystal Structure of DPPIV with a Chemibody Inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-{4-[(3R)-3-amino-4-(2,4,5-trifluorophenyl)butanoyl]piperazin-1-yl}-N-(22-oxo-3,6,9,12,15,18-hexaoxa-21-azatricosan-1-yl)acetamide, ... | | Authors: | Wang, Z, Johnstone, S, Cheng, A. | | Deposit date: | 2017-05-16 | | Release date: | 2018-05-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-guided Discovery of Dual-recognition Chemibodies.
Sci Rep, 8, 2018
|
|
6FL5
 
 | | Structure of human SHMT1-H135N-R137A-E168N mutant at 3.6 Ang. resolution | | Descriptor: | CHLORIDE ION, PYRIDOXAL-5'-PHOSPHATE, Serine hydroxymethyltransferase, ... | | Authors: | Giardina, G, Cutruzzola, F, Lucchi, R. | | Deposit date: | 2018-01-25 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | The catalytic activity of serine hydroxymethyltransferase is essential for de novo nuclear dTMP synthesis in lung cancer cells.
FEBS J., 285, 2018
|
|
7UB0
 
 | |
8DC9
 
 | | RNA ligase RtcB from Pyrococcus horikoshii in complex with Mn2+ and GTP | | Descriptor: | CHLORIDE ION, GUANOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Jacewicz, A, Dantuluri, S, Shuman, S. | | Deposit date: | 2022-06-16 | | Release date: | 2022-10-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Structures of RNA ligase RtcB in complexes with divalent cations and GTP.
Rna, 28, 2022
|
|
7UB6
 
 | |
7UAP
 
 | |
5JSE
 
 | | Crystal structure of phiAB6 tailspike in complex with three-repeated oligosaccharides of Acinetobacter baumannii surface polysaccharide | | Descriptor: | ACETIC ACID, MALONIC ACID, beta-D-galactopyranose-(1-3)-2-amino-2-deoxy-beta-D-galactopyranose-(1-3)-[5,7-bisacetamido-3,5,7,9-tetradeoxy-L-glycero-alpha-L-manno-non-2-ulopyranosonic acid-(2-6)-beta-D-glucopyranose-(1-6)]beta-D-galactopyranose-(1-3)-2-amino-2-deoxy-beta-D-galactopyranose-(1-3)-[beta-D-glucopyranose-(1-6)]beta-D-galactopyranose, ... | | Authors: | Lee, I.M, Tu, I.F, Huang, K.F, Wu, S.H. | | Deposit date: | 2016-05-08 | | Release date: | 2017-03-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural basis for fragmenting the exopolysaccharide of Acinetobacter baumannii by bacteriophage Phi AB6 tailspike protein
Sci Rep, 7, 2017
|
|
7UB5
 
 | |
8DCG
 
 | | Structure of guanylylated RNA ligase RtcB from Pyrococcus horikoshii | | Descriptor: | CHLORIDE ION, GUANOSINE-5'-MONOPHOSPHATE, SULFATE ION, ... | | Authors: | Jacewicz, A, Dantuluri, S, Shuman, S. | | Deposit date: | 2022-06-16 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structures of RNA ligase RtcB in complexes with divalent cations and GTP.
Rna, 28, 2022
|
|
6FIO
 
 | | DDR1, 2-[1'-(1H-indazole-5-carbonyl)-4-methyl-2-oxospiro[indole-3,4'-piperidine]-1-yl]-N-(2,2,2-trifluoroethyl)acetamide, 1.990A, P6522, Rfree=27.7% | | Descriptor: | 2-[1'-(1~{H}-indazol-5-ylcarbonyl)-4-methyl-2-oxidanylidene-spiro[indole-3,4'-piperidine]-1-yl]-~{N}-[2,2,2-tris(fluoranyl)ethyl]ethanamide, Epithelial discoidin domain-containing receptor 1 | | Authors: | Stihle, M, Richter, H, Benz, J, Kuhn, B, Rudolph, M.G. | | Deposit date: | 2018-01-19 | | Release date: | 2018-11-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | DNA-Encoded Library-Derived DDR1 Inhibitor Prevents Fibrosis and Renal Function Loss in a Genetic Mouse Model of Alport Syndrome.
Acs Chem.Biol., 14, 2019
|
|
7UAR
 
 | |
