3QLH
 
 | | HIV-1 Reverse Transcriptase in Complex with Manicol at the RNase H Active Site and TMC278 (Rilpivirine) at the NNRTI Binding Pocket | | Descriptor: | (2S)-5,7-dihydroxy-9-methyl-2-(prop-1-en-2-yl)-1,2,3,4-tetrahydro-6H-benzo[7]annulen-6-one, 1,2-ETHANEDIOL, 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, ... | | Authors: | Himmel, D.M, Wojtak, K, Bauman, J.D, Arnold, E. | | Deposit date: | 2011-02-02 | | Release date: | 2011-12-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Synthesis, activity, and structural analysis of novel alpha-hydroxytropolone inhibitors of human immunodeficiency virus reverse transcriptase-associated ribonuclease H.
J.Med.Chem., 54, 2011
|
|
2PVL
 
 | | Structure-Based Design of Pyrazolo[1,5-a][1,3,5]triazine Derivatives as Potent Inhibitors of Protein Kinase CK2 | | Descriptor: | 2-(4-ETHYLPIPERAZIN-1-YL)-4-(PHENYLAMINO)PYRAZOLO[1,5-A][1,3,5]TRIAZINE-8-CARBONITRILE, Casein kinase II subunit alpha | | Authors: | Nie, Z, Perretta, C, Erickson, P, Margosiak, S, Almassy, R, Lu, J, Averill, A, Yager, K.M, Chu, S. | | Deposit date: | 2007-05-09 | | Release date: | 2008-05-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-based design, synthesis, and study of pyrazolo[1,5-a][1,3,5]triazine derivatives as potent inhibitors of protein kinase CK2.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
3VF7
 
 | | Crystal Structure of HIV-1 Protease Mutant L76V with novel P1'-Ligands GRL-02031 | | Descriptor: | (3aS,5R,6aR)-hexahydro-2H-cyclopenta[b]furan-5-yl [(1S,2R)-1-benzyl-2-hydroxy-3-([(4-methoxyphenyl)sulfonyl]{[(2R)-5-oxopyrrolidin-2-yl]methyl}amino)propyl]carbamate, CHLORIDE ION, GLYCEROL, ... | | Authors: | Yu, X.X, Wang, Y.F, Chang, Y.C.E, Weber, I.T. | | Deposit date: | 2012-01-09 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Potent antiviral HIV-1 protease inhibitor GRL-02031 adapts to the structures of drug resistant mutants with its P1'-pyrrolidinone ring.
J.Med.Chem., 55, 2012
|
|
3VZD
 
 | | Crystal structure of Sphingosine Kinase 1 with inhibitor and ADP | | Descriptor: | 4-{[4-(4-chlorophenyl)-1,3-thiazol-2-yl]amino}phenol, ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Min, X, Walker, N.P, Wang, Z. | | Deposit date: | 2012-10-11 | | Release date: | 2013-05-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular basis of sphingosine kinase 1 substrate recognition and catalysis.
Structure, 21, 2013
|
|
1BOT
 
 | | CRYSTAL STRUCTURE OF THE COMPLEX BETWEEN ESCHERICHIA COLI GLYCEROL KINASE AND THE ALLOSTERIC REGULATOR FRUCTOSE 1,6-BISPHOSPHATE. | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, PROTEIN (GLYCEROL KINASE) | | Authors: | Ormo, M, Bystrom, C.E, Remington, S.J. | | Deposit date: | 1998-08-05 | | Release date: | 1999-01-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Crystal structure of a complex of Escherichia coli glycerol kinase and an allosteric effector fructose 1,6-bisphosphate.
Biochemistry, 37, 1998
|
|
4OF6
 
 | | Crystal Structure of SYG-1 D1, Crystal form 1 | | Descriptor: | 1,2-ETHANEDIOL, Protein SYG-1, isoform b, ... | | Authors: | Ozkan, E, Garcia, K.C. | | Deposit date: | 2014-01-14 | | Release date: | 2014-02-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.696 Å) | | Cite: | Extracellular Architecture of the SYG-1/SYG-2 Adhesion Complex Instructs Synaptogenesis.
Cell(Cambridge,Mass.), 156, 2014
|
|
1GX0
 
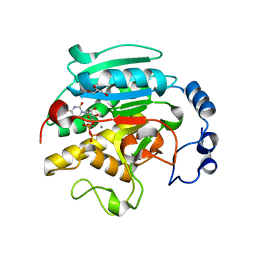 | | ALPHA-,1,3 GALACTOSYLTRANSFERASE - BETA-D-GALACTOSE COMPLEX | | Descriptor: | GLYCEROL, MANGANESE (II) ION, N-ACETYLLACTOSAMINIDE ALPHA-1,3-GALACTOSYLTRANSFERASE, ... | | Authors: | Boix, E, Zhang, Y, Swaminathan, G.J, Brew, K, Acharya, K.R. | | Deposit date: | 2002-03-26 | | Release date: | 2003-03-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of Ordered Binding of Donor and Acceptor Substrates to the Retaining Glycosyltransferase, Alpha -1,3 Galactosyltransferase
J.Biol.Chem., 277, 2002
|
|
4HE9
 
 | | Crystal Structure of HIV-1 protease mutants I54M complexed with inhibitor GRL-0519 | | Descriptor: | (3R,3aS,3bR,6aS,7aS)-octahydrodifuro[2,3-b:3',2'-d]furan-3-yl [(1S,2R)-1-benzyl-2-hydroxy-3-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}propyl]carbamate, CHLORIDE ION, HIV-1 protease, ... | | Authors: | Shen, C.H, Zhang, H, Weber, I.T. | | Deposit date: | 2012-10-03 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Novel P2 tris-tetrahydrofuran group in antiviral compound 1 (GRL-0519) fills the S2 binding pocket of selected mutants of HIV-1 protease.
J.Med.Chem., 56, 2013
|
|
1GZL
 
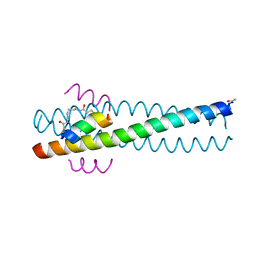 | | Crystal structure of C14linkmid/IQN17: a cross-linked inhibitor of HIV-1 entry bound to the gp41 hydrophobic pocket | | Descriptor: | CHLORIDE ION, ENVELOPE GLYCOPROTEIN GP41, FUSION PROTEIN BETWEEN THE HYDROPHOBIC POCKET OF HIV GP41 AND GENERAL CONTROL PROTEIN GCN4-PIQI, ... | | Authors: | Sia, S.K, Carr, P.A, Cochran, A.G, Malashkevich, V.M, Kim, P.S. | | Deposit date: | 2002-05-23 | | Release date: | 2002-10-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Short Constrained Peptides that Inhibit HIV-1 Entry
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
3VFB
 
 | | Crystal Structure of HIV-1 Protease Mutant N88D with novel P1'-Ligands GRL-02031 | | Descriptor: | (3aS,5R,6aR)-hexahydro-2H-cyclopenta[b]furan-5-yl [(1S,2R)-1-benzyl-2-hydroxy-3-([(4-methoxyphenyl)sulfonyl]{[(2R)-5-oxopyrrolidin-2-yl]methyl}amino)propyl]carbamate, CHLORIDE ION, GLYCEROL, ... | | Authors: | Yu, X.X, Wang, Y.F, Chang, Y.C.E, Weber, I.T. | | Deposit date: | 2012-01-09 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Potent antiviral HIV-1 protease inhibitor GRL-02031 adapts to the structures of drug resistant mutants with its P1'-pyrrolidinone ring.
J.Med.Chem., 55, 2012
|
|
2EGH
 
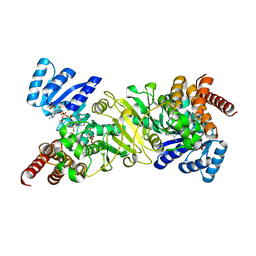 | | Crystal structure of 1-deoxy-D-xylulose 5-phosphate reductoisomerase complexed with a magnesium ion, NADPH and fosmidomycin | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, 3-[FORMYL(HYDROXY)AMINO]PROPYLPHOSPHONIC ACID, MAGNESIUM ION, ... | | Authors: | Yajima, S, Hara, K, Iino, D, Sasaki, Y, Kuzuyama, T, Seto, H. | | Deposit date: | 2007-03-01 | | Release date: | 2007-06-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of 1-deoxy-D-xylulose 5-phosphate reductoisomerase in a quaternary complex with a magnesium ion, NADPH and the antimalarial drug fosmidomycin
Acta Crystallogr.,Sect.F, 63, 2007
|
|
2AOC
 
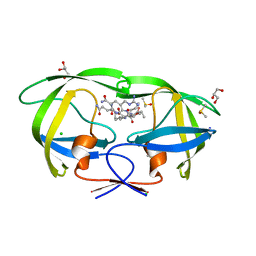 | | Crystal structure analysis of HIV-1 protease mutant I84V with a substrate analog P2-NC | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Tie, Y, Boross, P.I, Wang, Y.F, Gaddis, L, Liu, F, Chen, X, Tozser, J, Harrison, R.W, Weber, I.T. | | Deposit date: | 2005-08-12 | | Release date: | 2006-01-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Molecular basis for substrate recognition and drug resistance from 1.1 to 1.6 angstroms resolution crystal structures of HIV-1 protease mutants with substrate analogs.
Febs J., 272, 2005
|
|
3NF7
 
 | | Structural basis for a new mechanism of inhibition of HIV integrase identified by fragment screening and structure based design | | Descriptor: | 1,2-ETHANEDIOL, 5-[(5-chloro-2-oxo-2,3-dihydro-1H-indol-1-yl)methyl]-1,3-benzodioxole-4-carboxylic acid, ACETIC ACID, ... | | Authors: | Peat, T.S, Newman, J, Deadman, J.J, Rhodes, D. | | Deposit date: | 2010-06-09 | | Release date: | 2011-04-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for a new mechanism of inhibition of HIV-1 integrase identified by fragment screening and structure-based design
ANTIVIR.CHEM.CHEMOTHER., 21, 2011
|
|
3NF6
 
 | | Structural basis for a new mechanism of inhibition of HIV integrase identified by fragment screening and structure based design | | Descriptor: | 1,2-ETHANEDIOL, 5-[(2-oxo-2,3-dihydro-1H-indol-1-yl)methyl]-1,3-benzodioxole-4-carboxylic acid, ACETIC ACID, ... | | Authors: | Peat, T.S, Newman, J, Deadman, J.J, Rhodes, D. | | Deposit date: | 2010-06-09 | | Release date: | 2011-04-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for a new mechanism of inhibition of HIV-1 integrase identified by fragment screening and structure-based design
ANTIVIR.CHEM.CHEMOTHER., 21, 2011
|
|
1H5R
 
 | | Thymidylyltransferase complexed with Thimidine and Glucose-1-phospate | | Descriptor: | 1-O-phosphono-alpha-D-glucopyranose, GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE, SULFATE ION, ... | | Authors: | Rosano, C, Zuccotti, S, Bolognesi, M. | | Deposit date: | 2001-05-25 | | Release date: | 2001-11-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Kinetic and Crystallographic Analyses Support a Sequential-Ordered Bi Bi Catalytic Mechanism for Escherichia Coli Glucose-1-Phosphate Thymidylyltransferase
J.Mol.Biol., 313, 2001
|
|
1RCO
 
 | | SPINACH RUBISCO IN COMPLEX WITH THE INHIBITOR D-XYLULOSE-2,2-DIOL-1,5-BISPHOSPHATE | | Descriptor: | D-XYLULOSE-2,2-DIOL-1,5-BISPHOSPHATE, RIBULOSE BISPHOSPHATE CARBOXYLASE/OXYGENASE | | Authors: | Taylor, T.C, Andersson, I. | | Deposit date: | 1996-10-31 | | Release date: | 1997-03-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A common structural basis for the inhibition of ribulose 1,5-bisphosphate carboxylase by 4-carboxyarabinitol 1,5-bisphosphate and xylulose 1,5-bisphosphate.
J.Biol.Chem., 271, 1996
|
|
1HBV
 
 | |
3NF9
 
 | | Structural basis for a new mechanism of inhibition of HIV integrase identified by fragment screening and structure based design | | Descriptor: | 1,2-ETHANEDIOL, 5-[(6-chloro-2-oxo-2,3-dihydro-1H-indol-1-yl)methyl]-1,3-benzodioxole-4-carboxylic acid, ACETIC ACID, ... | | Authors: | Peat, T.S, Newman, J, Deadman, J.J, Rhodes, D. | | Deposit date: | 2010-06-10 | | Release date: | 2011-04-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for a new mechanism of inhibition of HIV-1 integrase identified by fragment screening and structure-based design
ANTIVIR.CHEM.CHEMOTHER., 21, 2011
|
|
3DK1
 
 | | Wild Type HIV-1 Protease with potent Antiviral inhibitor GRL-0105A | | Descriptor: | (4aR,6r,7aS)-hexahydro-4aH-cyclopenta[b][1,4]dioxin-6-yl [(1S,2R)-1-benzyl-2-hydroxy-3-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}propyl]carbamate, CHLORIDE ION, Protease, ... | | Authors: | Wang, Y.F, Weber, I.T. | | Deposit date: | 2008-06-24 | | Release date: | 2009-05-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Potent HIV-1 protease inhibitors incorporating meso-bicyclic urethanes as P2-ligands: structure-based design, synthesis, biological evaluation and protein-ligand X-ray studies
Org.Biomol.Chem., 6, 2008
|
|
2AZ9
 
 | | HIV-1 Protease NL4-3 1X mutant | | Descriptor: | PROTEASE RETROPEPSIN, benzyl [(1S,4S,7S,8R,9R,10S,13S,16S)-7,10-dibenzyl-8,9-dihydroxy-1,16-dimethyl-4,13-bis(1-methylethyl)-2,5,12,15,18-pentaoxo-20-phenyl-19-oxa-3,6,11,14,17-pentaazaicos-1-yl]carbamate | | Authors: | Heaslet, H, Kutilek, V, Morris, G.M, Lin, Y.-C, Elder, J.H, Torbett, B.E, Stout, C.D. | | Deposit date: | 2005-09-09 | | Release date: | 2006-02-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Insights into the Mechanisms of Drug Resistance in HIV-1 Protease NL4-3
J.Mol.Biol., 356, 2006
|
|
2AOG
 
 | | Crystal structure analysis of HIV-1 protease mutant V82A with a substrate analog P2-NC | | Descriptor: | ACETIC ACID, GLYCEROL, HIV-1 PROTEASE (RETROPEPSIN), ... | | Authors: | Tie, Y, Boross, P.I, Wang, Y.F, Gaddis, L, Liu, F, Chen, X, Tozser, J, Harrison, R.W, Weber, I.T. | | Deposit date: | 2005-08-12 | | Release date: | 2006-01-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Molecular basis for substrate recognition and drug resistance from 1.1 to 1.6 angstroms resolution crystal structures of HIV-1 protease mutants with substrate analogs.
Febs J., 272, 2005
|
|
2AOD
 
 | | Crystal structure analysis of HIV-1 protease with a substrate analog P2-NC | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, HIV-1 PROTEASE, ... | | Authors: | Tie, Y, Boross, P.I, Wang, Y.F, Gaddis, L, Liu, F, Chen, X, Tozser, J, Harrison, R.W, Weber, I.T. | | Deposit date: | 2005-08-12 | | Release date: | 2006-01-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Molecular basis for substrate recognition and drug resistance from 1.1 to 1.6 angstroms resolution crystal structures of HIV-1 protease mutants with substrate analogs.
Febs J., 272, 2005
|
|
2VS4
 
 | | THE BINDING OF UDP-GALACTOSE BY AN ACTIVE SITE MUTANT OF alpha-1,3 GALACTOSYLTRANSFERASE (alpha3GT) | | Descriptor: | GLYCEROL, MANGANESE (II) ION, N-ACETYLLACTOSAMINIDE ALPHA-1,3-GALACTOSYLTRANSFERASE, ... | | Authors: | Tumbale, P, Jamaluddin, H, Thiyagarajan, N, Brew, K, Acharya, K.R. | | Deposit date: | 2008-04-18 | | Release date: | 2008-07-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural Basis of Udp-Galactose Binding by Alpha- 1,3-Galactosyltransferase (Alpha3Gt): Role of Negative Charge on Aspartic Acid 316 in Structure and Activity.
Biochemistry, 47, 2008
|
|
2WZE
 
 | | High resolution crystallographic structure of the Clostridium thermocellum N-terminal endo-1,4-beta-D-xylanase 10B (Xyn10B) CBM22-1- GH10 modules complexed with xylohexaose | | Descriptor: | CALCIUM ION, ENDO-1,4-BETA-XYLANASE Y, GLYCEROL, ... | | Authors: | Najmudin, S, Pinheiro, B.A, Romao, M.J, Prates, J.A.M, Fontes, C.M.G.A. | | Deposit date: | 2009-11-27 | | Release date: | 2010-08-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Putting an N-Terminal End to the Clostridium Thermocellum Xylanase Xyn10B Story: Crystal Structure of the Cbm22-1-Gh10 Modules Complexed with Xylohexaose.
J.Struct.Biol., 172, 2010
|
|
1HFS
 
 | |
