5HFA
 
 | | Crystal structure of human acetylcholinesterase in complex with paraoxon and 2-PAM | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Franklin, M.F, Rudolph, M.J, Ginter, C, Cassidy, M.S, Cheung, J. | | Deposit date: | 2016-01-06 | | Release date: | 2016-06-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Structures of paraoxon-inhibited human acetylcholinesterase reveal perturbations of the acyl loop and the dimer interface.
Proteins, 84, 2016
|
|
5HET
 
 | |
5HFD
 
 | |
5ZIA
 
 | |
5ZLD
 
 | |
5HI7
 
 | | Co-crystal structure of human SMYD3 with an aza-SAH compound | | Descriptor: | 5'-{[(3S)-3-amino-3-carboxypropyl][3-(dimethylamino)propyl]amino}-5'-deoxyadenosine, DIMETHYL SULFOXIDE, Histone-lysine N-methyltransferase SMYD3, ... | | Authors: | Elkins, P.A, Bonnette, W.G. | | Deposit date: | 2016-01-11 | | Release date: | 2016-03-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure-Based Design of a Novel SMYD3 Inhibitor that Bridges the SAM-and MEKK2-Binding Pockets.
Structure, 24, 2016
|
|
7ZPV
 
 | | Room temperature SSX crystal structure of CTX-M-14 | | Descriptor: | Beta-lactamase, SULFATE ION | | Authors: | Oberthuer, D, Perbandt, M, Prester, A, Rohde, H, Betzel, C, Yefanov, O. | | Deposit date: | 2022-04-29 | | Release date: | 2022-11-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Rapid and efficient room-temperature serial synchrotron crystallography using the CFEL TapeDrive.
Iucrj, 9, 2022
|
|
5Z8I
 
 | |
5ZCZ
 
 | | Solution structure of the T. Thermophilus HB8 TTHA1718 protein in living eukaryotic cells by in-cell NMR spectroscopy | | Descriptor: | Heavy metal binding protein | | Authors: | Tanaka, T, Teppei, I, Kamoshida, H, Mishima, M, Shirakawa, M, Guentert, P, Ito, Y. | | Deposit date: | 2018-02-22 | | Release date: | 2019-08-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | High-Resolution Protein 3D Structure Determination in Living Eukaryotic Cells.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
5Z8Q
 
 | |
5Z4F
 
 | | An anthrahydroquino-Gama-pyrone synthase Txn09 complexed with SUM | | Descriptor: | 1,8-dihydroxy-2-[(4R)-4-hydroxy-4-methyl-3-oxohexanoyl]-3-methylanthracene-9,10-dione, TxnO9 | | Authors: | Song, Y.J, Cao, C.Y. | | Deposit date: | 2018-01-11 | | Release date: | 2019-01-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Enzymology of Anthraquinone-gamma-Pyrone Ring Formation in Complex Aromatic Polyketide Biosynthesis.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5ZD0
 
 | | Solution structure of human ubiquitin with three alanine mutations in living eukaryotic cells by in-cell NMR spectroscopy | | Descriptor: | ubiquitin | | Authors: | Tanaka, T, Ikeya, T, Kamoshida, H, Mishima, M, Shirakawa, M, Guentert, P, Ito, Y. | | Deposit date: | 2018-02-22 | | Release date: | 2019-08-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | High-Resolution Protein 3D Structure Determination in Living Eukaryotic Cells.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
5ZPV
 
 | |
5Z4E
 
 | | An anthrahydroquino-Gama-pyrone synthase Txn09 | | Descriptor: | TxnO9 | | Authors: | Song, Y.J, Cao, C.Y. | | Deposit date: | 2018-01-11 | | Release date: | 2018-12-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Enzymology of Anthraquinone-gamma-Pyrone Ring Formation in Complex Aromatic Polyketide Biosynthesis.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5Z9C
 
 | |
5ZMR
 
 | | Solution Structure of the N-terminal Domain of the Yeast Rpn5 | | Descriptor: | 26S proteasome regulatory subunit RPN5 | | Authors: | Zhang, W, Zhao, C, Li, H, Hu, Y, Jin, C. | | Deposit date: | 2018-04-05 | | Release date: | 2018-09-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal domain of proteasome lid subunit Rpn5
Biochem. Biophys. Res. Commun., 504, 2018
|
|
5YXI
 
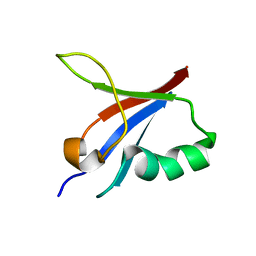 | | Designed protein dRafX6 | | Descriptor: | Designed protein dRafX6 | | Authors: | Liu, R. | | Deposit date: | 2017-12-05 | | Release date: | 2018-12-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | De novo sequence redesign of a functional Ras-binding domain globally inverted the surface charge distribution and led to extreme thermostability.
Biotechnol.Bioeng., 118, 2021
|
|
5HR1
 
 | | Crystal structure of thioredoxin L107A mutant | | Descriptor: | COPPER (II) ION, Thioredoxin-1 | | Authors: | Noguera, M.E, Vazquez, D.S, Howard, E.I, Cousido-Siah, A, Mitschler, A, Podjarny, A, Santos, J. | | Deposit date: | 2016-01-22 | | Release date: | 2017-02-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.144 Å) | | Cite: | Structural variability of E. coli thioredoxin captured in the crystal structures of single-point mutants.
Sci Rep, 7, 2017
|
|
5Z4B
 
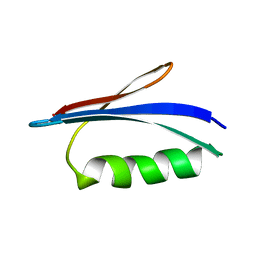 | | GB1 structure determination in living eukaryotic cells by in-cell NMR spectroscopy | | Descriptor: | Protein LG | | Authors: | Tanaka, T, Teppei, I, Kamoshida, H, Mishima, M, Shirakawa, M, Guentert, P, Ito, Y. | | Deposit date: | 2018-01-10 | | Release date: | 2019-01-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | High-Resolution Protein 3D Structure Determination in Living Eukaryotic Cells.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
8HT7
 
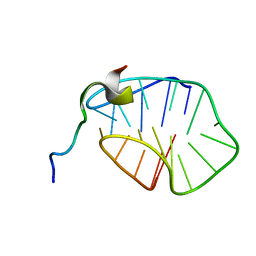 | | The N-terminal region of Cdc6 specifically recognizes human DNA G-quadruplex | | Descriptor: | DNA (5'-D(*GP*GP*GP*TP*TP*AP*GP*GP*GP*TP*TP*AP*GP*GP*GP*TP*TP*TP*GP*GP*G)-3'), GLN-ALA-GLN-ALA-THR-ILE-SER-PHE-PRO-LYS-ARG-LYS-LEU-SER-TRP | | Authors: | Liu, C, Zhu, G, Geng, Y, Xu, N. | | Deposit date: | 2022-12-20 | | Release date: | 2023-12-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The N-terminal region of Cdc6 specifically recognizes human DNA G-quadruplex.
Int.J.Biol.Macromol., 260, 2024
|
|
1X3F
 
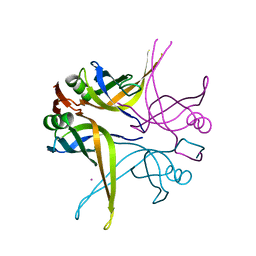 | | Crystal structure of the single-stranded DNA-binding protein from Mycobacterium SMEGMATIS | | Descriptor: | CADMIUM ION, Single-strand binding protein | | Authors: | Saikrishnan, K, Manjunath, G.P, Singh, P, Jeyakanthan, J, Dauter, Z, Sekar, K, Muniyappa, K, Vijayan, M. | | Deposit date: | 2005-05-05 | | Release date: | 2005-08-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of Mycobacterium smegmatis single-stranded DNA-binding protein and a comparative study involving homologus SSBs: biological implications of structural plasticity and variability in quaternary association.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2M4V
 
 | | Mycobacterium tuberculosis RNA polymerase binding protein A (RbpA) and its interactions with sigma factors | | Descriptor: | Putative uncharacterized protein | | Authors: | Bortoluzzi, A, Muskett, F.W, Waters, L.C, Addis, P.W, Rieck, B, Munder, T, Schleier, S, Forti, F, Ghisotti, D, Carr, M.D, O'Hare, H.M. | | Deposit date: | 2013-02-11 | | Release date: | 2013-04-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Mycobacterium tuberculosis RNA polymerase-binding protein A (RbpA) and its interactions with sigma factors.
J.Biol.Chem., 288, 2013
|
|
5HX2
 
 | | In vitro assembled star-shaped hubless T4 baseplate | | Descriptor: | Baseplate wedge protein gp10, Baseplate wedge protein gp53, Baseplate wedge protein gp6, ... | | Authors: | Yap, M.L, Klose, T, Fokine, A, Rossmann, M.G. | | Deposit date: | 2016-01-29 | | Release date: | 2016-03-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Role of bacteriophage T4 baseplate in regulating assembly and infection.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5HVT
 
 | | Crystal Structure of Macrophage Migration Inhibitory Factor (MIF) with a Potent Inhibitor (NVS-2) | | Descriptor: | 7-hydroxy-3-(4-methoxyphenyl)-3,4-dihydro-2H-1,3-benzoxazin-2-one, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Robertson, M.J, Jorgensen, W.L. | | Deposit date: | 2016-01-28 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A Fluorescence Polarization Assay for Binding to Macrophage Migration Inhibitory Factor and Crystal Structures for Complexes of Two Potent Inhibitors.
J.Am.Chem.Soc., 138, 2016
|
|
5HFB
 
 | |
