7TNF
 
 | |
7TNU
 
 | |
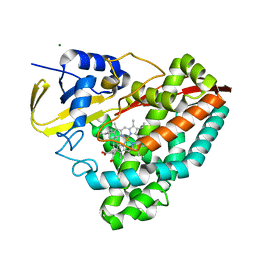
3V7U
 
 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with MTA | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, CITRIC ACID, GLYCEROL, ... | | Authors: | Gelin, M, Poncet-Montange, G, Assairi, L, Morellato, L, Huteau, V, Dugu, L, Dussurget, O, Pochet, S, Labesse, G. | | Deposit date: | 2011-12-22 | | Release date: | 2012-03-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Screening and In Situ Synthesis Using Crystals of a NAD Kinase Lead to a Potent Antistaphylococcal Compound.
Structure, 20, 2012
|
|
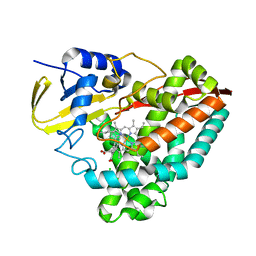
3V8N
 
 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with 8-bromo-5'-amino-5'-deoxyadenosine, reacted with a citrate molecule in N site | | Descriptor: | 5'-amino-8-bromo-5'-deoxyadenosine, 8-bromo-5'-{[3-carboxy-2-(carboxymethyl)-2-hydroxypropanoyl]amino}-5'-deoxyadenosine, CITRIC ACID, ... | | Authors: | Gelin, M, Poncet-Montange, G, Assairi, L, Morellato, L, Huteau, V, Dugu, L, Dussurget, O, Pochet, S, Labesse, G. | | Deposit date: | 2011-12-23 | | Release date: | 2012-03-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3801 Å) | | Cite: | Screening and In Situ Synthesis Using Crystals of a NAD Kinase Lead to a Potent Antistaphylococcal Compound.
Structure, 20, 2012
|
|
4FDN
 
 | |
3V8R
 
 | | Crystal structure of NAD kinase 1 H223E mutant from Listeria monocytogenes in complex with 5'-amino-8-bromo-5'-deoxyadenosine | | Descriptor: | 5'-amino-8-bromo-5'-deoxyadenosine, CITRIC ACID, Probable inorganic polyphosphate/ATP-NAD kinase 1 | | Authors: | Gelin, M, Poncet-Montange, G, Assairi, L, Morellato, L, Huteau, V, Dugu, L, Dussurget, O, Pochet, S, Labesse, G. | | Deposit date: | 2011-12-23 | | Release date: | 2012-03-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Screening and In Situ Synthesis Using Crystals of a NAD Kinase Lead to a Potent Antistaphylococcal Compound.
Structure, 20, 2012
|
|
5IPR
 
 | | Cryo-EM structure of GluN1/GluN2B NMDA receptor in the DCKA/D-APV-bound conformation, state 3 | | Descriptor: | Ionotropic glutamate receptor subunit NR2B, N-methyl-D-aspartate receptor subunit NR1-8a | | Authors: | Zhu, S, Stein, A.R, Yoshioka, C, Lee, C.H, Goehring, A, Mchaourab, S.H, Gouaux, E. | | Deposit date: | 2016-03-09 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (14.1 Å) | | Cite: | Mechanism of NMDA Receptor Inhibition and Activation.
Cell, 165, 2016
|
|
5ZB2
 
 | | Crystal structure of Rad7 and Elc1 complex in yeast | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 3,6,9,12,15,18,21,24,27,30,33,36,39,42,45,48,51,54,57-nonadecaoxanonapentacontane-1,59-diol, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, ... | | Authors: | Jiang, T, Liu, L, Huo, Y. | | Deposit date: | 2018-02-09 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.30000544 Å) | | Cite: | Crystal structure of the yeast Rad7-Elc1 complex and assembly of the Rad7-Rad16-Elc1-Cul3 complex.
DNA Repair (Amst.), 77, 2019
|
|
5IPQ
 
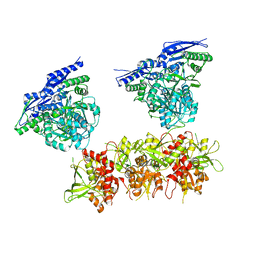 | | Cryo-EM structure of GluN1/GluN2B NMDA receptor in the DCKA/D-APV-bound conformation, state 2 | | Descriptor: | Ionotropic glutamate receptor subunit NR2B, N-methyl-D-aspartate receptor subunit NR1-8a | | Authors: | Zhu, S, Stein, A.R, Yoshioka, C, Lee, C.H, Goehring, A, Mchaourab, S.H, Gouaux, E. | | Deposit date: | 2016-03-09 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (13.5 Å) | | Cite: | Mechanism of NMDA Receptor Inhibition and Activation.
Cell, 165, 2016
|
|
5W9S
 
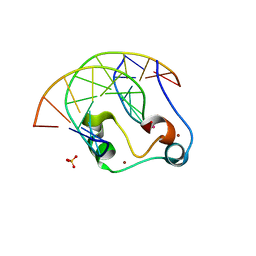 | | Zinc finger of human CXXC5 in complex with CpG DNA | | Descriptor: | CXXC-type zinc finger protein 5, CpG DNA fragment, SULFATE ION, ... | | Authors: | Liu, K, Xu, C, Tempel, W, Walker, J.R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-06-23 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | DNA Sequence Recognition of Human CXXC Domains and Their Structural Determinants.
Structure, 26, 2018
|
|
3I6A
 
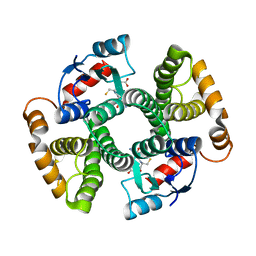 | | Human GST A1-1 GIMF mutant with Glutathione | | Descriptor: | GLUTATHIONE, Glutathione S-transferase A1 | | Authors: | Balogh, L.M, Le Trong, I, Stenkamp, R.E, Atkins, W.M. | | Deposit date: | 2009-07-06 | | Release date: | 2009-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural analysis of a glutathione transferase A1-1 mutant tailored for high catalytic efficiency with toxic alkenals.
Biochemistry, 48, 2009
|
|
2IVV
 
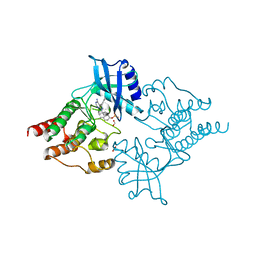 | | Crystal structure of phosphorylated RET tyrosine kinase domain complexed with the inhibitor PP1 | | Descriptor: | 1-TER-BUTYL-3-P-TOLYL-1H-PYRAZOLO[3,4-D]PYRIMIDIN-4-YLAMINE, FORMIC ACID, PROTO-ONCOGENE TYROSINE-PROTEIN KINASE RECEPTOR RET PRECURSOR | | Authors: | Knowles, P.P, Murray-Rust, J, McDonald, N.Q. | | Deposit date: | 2006-06-16 | | Release date: | 2006-08-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure and chemical inhibition of the RET tyrosine kinase domain.
J. Biol. Chem., 281, 2006
|
|
4FWU
 
 | | Crystal structure of glutaminyl cyclase from drosophila melanogaster in space group I4 | | Descriptor: | 1,2-ETHANEDIOL, CG32412, SULFATE ION, ... | | Authors: | Kolenko, P, Koch, B, Stubbs, M.T. | | Deposit date: | 2012-07-02 | | Release date: | 2012-09-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of glutaminyl cyclase from Drosophila melanogaster in space group I4.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
5IOU
 
 | | Cryo-EM structure of GluN1/GluN2B NMDA receptor in the glutamate/glycine-bound conformation | | Descriptor: | GLUTAMIC ACID, GLYCINE, Ionotropic glutamate receptor subunit NR2B, ... | | Authors: | Zhu, S, Stein, A.R, Yoshioka, C, Lee, C.H, Goehring, A, Mchaourab, S.H, Gouaux, E. | | Deposit date: | 2016-03-09 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Mechanism of NMDA Receptor Inhibition and Activation.
Cell, 165, 2016
|
|
5IPU
 
 | | Cryo-EM structure of GluN1/GluN2B NMDA receptor in the DCKA/D-APV-bound conformation, state 6 | | Descriptor: | Ionotropic glutamate receptor subunit NR2B, N-methyl-D-aspartate receptor subunit NR1-8a | | Authors: | Zhu, S, Stein, A.R, Yoshioka, C, Lee, C.H, Goehring, A, Mchaourab, S.H, Gouaux, E. | | Deposit date: | 2016-03-09 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (15.4 Å) | | Cite: | Mechanism of NMDA Receptor Inhibition and Activation.
Cell, 165, 2016
|
|
4FSL
 
 | |
1VEB
 
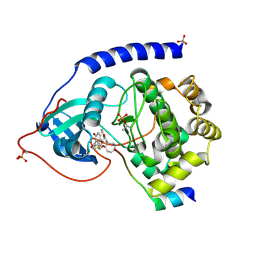 | | Crystal Structure of Protein Kinase A in Complex with Azepane Derivative 5 | | Descriptor: | (3R,4R)-N-{4-[4-(2-FLUORO-6-HYDROXY-3-METHOXY-BENZOYL)-BENZYLOXY]-AZEPAN-3-YL}-ISONICOTINAMIDE, cAMP-dependent protein kinase inhibitor, alpha form, ... | | Authors: | Breitenlechner, C.B, Wegge, T, Berillon, L, Graul, K, Marzenell, K, Friebe, W.-G, Thomas, U, Schumacher, R, Huber, R, Engh, R.A, Masjost, B. | | Deposit date: | 2004-03-29 | | Release date: | 2005-03-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structure-based optimization of novel azepane derivatives as PKB inhibitors
J.Med.Chem., 47, 2004
|
|
4BLC
 
 | | THE STRUCTURE OF ORTHORHOMBIC CRYSTALS OF BEEF LIVER CATALASE | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROTEIN (CATALASE), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Ko, T.P, Day, J, Malkin, A, McPherson, A. | | Deposit date: | 1998-09-27 | | Release date: | 1998-10-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of orthorhombic crystals of beef liver catalase.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
2ZTI
 
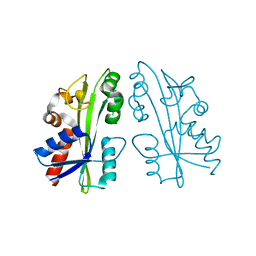 | |
2FPG
 
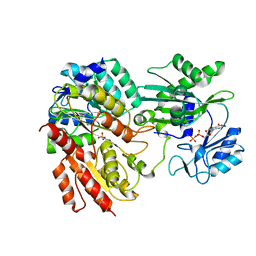 | |
8IJN
 
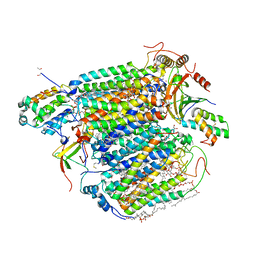 | | Bovine Heart Cytochrome c Oxidase in the Nitric Oxide-Bound Fully Reduced State at 100 K | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Tsukihara, T, Shimada, A, Muramoto, K. | | Deposit date: | 2023-02-27 | | Release date: | 2023-04-12 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Bovine cytochrome c oxidase structures enable O2 reduction with minimization of reactive oxygens and provide a proton-pumping gate.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
7L1A
 
 | | Human Methionine Adenosyltransferase 2A bound to Methylthioadenosine and inhibitor, di-imido triphosphate (PNPNP) | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, ALANINE, MAGNESIUM ION, ... | | Authors: | Niland, C.N, Fedorov, E, Schramm, V.L, Ghosh, A. | | Deposit date: | 2020-12-14 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Mechanism and Inhibition of Human Methionine Adenosyltransferase 2A.
Biochemistry, 60, 2021
|
|
5T71
 
 | | Human carboanhydrase F131C_C206S double mutant in complex with SA-2 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(HYDROXYMERCURY)BENZOIC ACID, 4-[(E)-diazenyl]benzene-1-sulfonamide, ... | | Authors: | DuBay, K.H, Iwan, K, Osorio-Planes, L, Geissler, P, Groll, M, Trauner, D, Broichhagen, J. | | Deposit date: | 2016-09-02 | | Release date: | 2017-09-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | A Predictive Approach for the Optical Control of Carbonic Anhydrase II Activity.
ACS Chem. Biol., 13, 2018
|
|
3LGW
 
 | |
7TXF
 
 | |
