6IWB
 
 | | Crystal structure of a computationally designed protein (LD3) in complex with BCL-2 | | Descriptor: | Apolipoprotein E, Apoptosis regulator Bcl-2,Apoptosis regulator Bcl-2, SULFATE ION | | Authors: | Kim, S, Kwak, M.J, Oh, B.-H, Correia, B.E, Gainza, P. | | Deposit date: | 2018-12-05 | | Release date: | 2019-12-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A computationally designed chimeric antigen receptor provides a small-molecule safety switch for T-cell therapy.
Nat.Biotechnol., 38, 2020
|
|
5KTG
 
 | |
8IQK
 
 | | Structural basis of the specificity and interaction mechanism of Bmf binding to pro-survival proteins | | Descriptor: | Bcl-2-like protein 1, Bcl-2-modifying factor | | Authors: | Wang, H, Guo, M, Wei, H, Chen, Y. | | Deposit date: | 2023-03-16 | | Release date: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.879 Å) | | Cite: | Structural basis of the specificity and interaction mechanism of Bmf binding to pro-survival Bcl-2 family proteins.
Comput Struct Biotechnol J, 21, 2023
|
|
8IQL
 
 | | Structural basis of the specificity and interaction mechanism of Bmf binding to pro-survival proteins | | Descriptor: | Apoptosis regulator Bcl-2, Bcl-2-modifying factor | | Authors: | Wang, H, Guo, M, Wei, H, Chen, Y. | | Deposit date: | 2023-03-16 | | Release date: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9577 Å) | | Cite: | Structural basis of the specificity and interaction mechanism of Bmf binding to pro-survival Bcl-2 family proteins.
Comput Struct Biotechnol J, 21, 2023
|
|
3R85
 
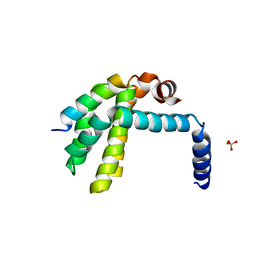 | | Crystal structure of human SOUL BH3 domain in complex with Bcl-xL | | Descriptor: | Bcl-2-like protein 1, Heme-binding protein 2, SULFATE ION | | Authors: | Ambrosi, E.K, Capaldi, S, Bovi, M, Saccomani, G, Perduca, M, Monaco, H.L. | | Deposit date: | 2011-03-23 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural changes in the BH3 domain of SOUL protein upon interaction with the anti-apoptotic protein Bcl-xL.
Biochem.J., 438, 2011
|
|
8EL0
 
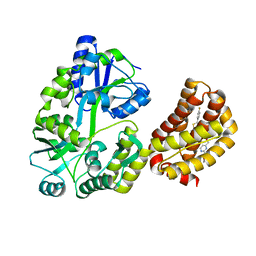 | | Structure of MBP-Mcl-1 in complex with a macrocyclic compound | | Descriptor: | (7R,20P)-18-chloro-1-(4-fluorophenyl)-10-{[(2M)-2-(2-methoxyphenyl)pyrimidin-4-yl]methoxy}-19-methyl-15-[2-(4-methylpiperazin-1-yl)ethyl]-7,8,15,16-tetrahydro-14H-17,20-etheno-9,13-(metheno)-6-oxa-2-thia-3,5,15-triazacyclooctadeca[1,2,3-cd]indene-7-carboxylic acid, Maltose/maltodextrin-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Judge, R.A, Judd, A.S, Souers, A.J. | | Deposit date: | 2022-09-22 | | Release date: | 2023-10-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.917 Å) | | Cite: | Selective MCL-1 inhibitor ABBV-467 is efficacious in tumor models but is associated with cardiac troponin increases in patients.
Commun Med (Lond), 3, 2023
|
|
8EL1
 
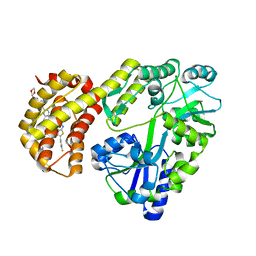 | | Structure of MBP-Mcl-1 in complex with ABBV-467 | | Descriptor: | (7R,16R)-19,23-dichloro-10-{[2-(4-{[(2R)-1,4-dioxan-2-yl]methoxy}phenyl)pyrimidin-4-yl]methoxy}-1-(4-fluorophenyl)-20,22-dimethyl-16-[(4-methylpiperazin-1-yl)methyl]-7,8,15,16-tetrahydro-18,21-etheno-13,9-(metheno)-6,14,17-trioxa-2-thia-3,5-diazacyclononadeca[1,2,3-cd]indene-7-carboxylic acid, Maltose/maltodextrin-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Judge, R.A, Judd, A.S, Souers, A.J. | | Deposit date: | 2022-09-22 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.406 Å) | | Cite: | Selective MCL-1 inhibitor ABBV-467 is efficacious in tumor models but is associated with cardiac troponin increases in patients.
Commun Med (Lond), 3, 2023
|
|
1LXL
 
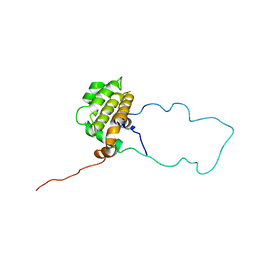 | | NMR STRUCTURE OF BCL-XL, AN INHIBITOR OF PROGRAMMED CELL DEATH, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | BCL-XL | | Authors: | Muchmore, S.W, Sattler, M, Liang, H, Meadows, R.P, Harlan, J.E, Yoon, H.S, Nettesheim, D, Chang, B.S, Thompson, C.B, Wong, S.L, Ng, S.C, Fesik, S.W. | | Deposit date: | 1996-04-04 | | Release date: | 1997-04-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | X-ray and NMR structure of human Bcl-xL, an inhibitor of programmed cell death.
Nature, 381, 1996
|
|
3QKD
 
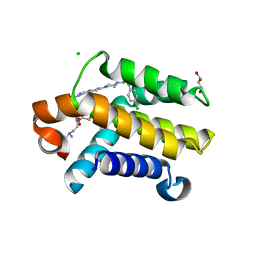 | | Crystal structure of Bcl-xL in complex with a Quinazoline sulfonamide inhibitor | | Descriptor: | (R)-N-(7-(4-((4'-chlorobiphenyl-2-yl)methyl)piperazin-1-yl)quinazolin-4-yl)-4-(4-(dimethylamino)-1-(phenylthio)butan-2-ylamino)-3-nitrobenzenesulfonamide, Bcl-2-like protein 1, CHLORIDE ION, ... | | Authors: | Czabotar, P.E, Smith, B.J. | | Deposit date: | 2011-01-31 | | Release date: | 2011-04-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Quinazoline sulfonamides as dual binders of the proteins B-cell lymphoma 2 and B-cell lymphoma extra long with potent proapoptotic cell-based activity.
J.Med.Chem., 54, 2011
|
|
7LHB
 
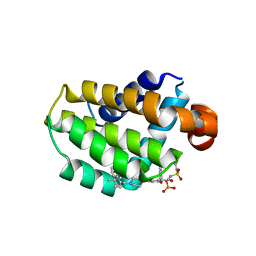 | | Crystal structure of Bcl-2 in complex with prodrug ABBV-167 | | Descriptor: | Apoptosis regulator Bcl-2, Phosphoric acid mono-[5-(5-{4-[2-(4-chloro-phenyl)-4,4-dimethyl-cyclohex-1-enylmethyl]-piperazin-1-yl}-2-{3-nitro-4-[(tetrahydro-pyran-4-ylmethyl)-amino]-benzenesulfonylaminocarbonyl}-phenoxy)-pyrrolo[2,3-b]pyridin-7-ylmethyl] ester | | Authors: | Judge, R.A, Salem, A.H. | | Deposit date: | 2021-01-21 | | Release date: | 2021-04-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.068 Å) | | Cite: | Expanding the Repertoire for "Large Small Molecules": Prodrug ABBV-167 Efficiently Converts to Venetoclax with Reduced Food Effect in Healthy Volunteers.
Mol.Cancer Ther., 20, 2021
|
|
1O0L
 
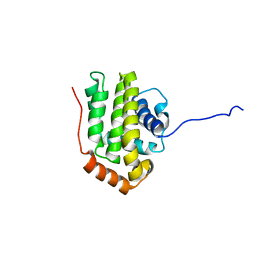 | | THE STRUCTURE OF BCL-W REVEALS A ROLE FOR THE C-TERMINAL RESIDUES IN MODULATING BIOLOGICAL ACTIVITY | | Descriptor: | Apoptosis regulator Bcl-W | | Authors: | Hinds, M.G, Lackmann, M, Skea, G.L, Harrison, P.J, Huang, D.C.S, Day, C.L. | | Deposit date: | 2003-02-22 | | Release date: | 2003-04-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure of Bcl-w reveals a role for the C-terminal residues in modulating biological activity
Embo J., 22, 2003
|
|
1OHU
 
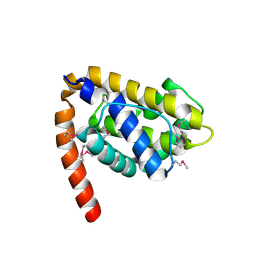 | | Structure of Caenorhabditis elegans CED-9 | | Descriptor: | APOPTOSIS REGULATOR CED-9 | | Authors: | Jeong, J.-S, Ha, N.-C, Oh, B.-H. | | Deposit date: | 2003-05-31 | | Release date: | 2003-08-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Unique Structural Features of a Bcl-2 Family Protein Ced-9 and Biophysical Characterization of Ced-9/Egl-1 Interactions
Cell Death Differ., 10, 2003
|
|
3KJ0
 
 | | Mcl-1 in complex with Bim BH3 mutant I2dY | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Bcl-2-like protein 11, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Fire, E, Grant, R.A, Keating, A.E. | | Deposit date: | 2009-11-02 | | Release date: | 2010-02-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mcl-1-Bim complexes accommodate surprising point mutations via minor structural changes.
Protein Sci., 19, 2010
|
|
3KJ1
 
 | | Mcl-1 in complex with Bim BH3 mutant I2dA | | Descriptor: | ACETATE ION, Bcl-2-like protein 11, CHLORIDE ION, ... | | Authors: | Fire, E, Grant, R.A, Keating, A.E. | | Deposit date: | 2009-11-02 | | Release date: | 2010-02-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.945 Å) | | Cite: | Mcl-1-Bim complexes accommodate surprising point mutations via minor structural changes.
Protein Sci., 19, 2010
|
|
3KJ2
 
 | | Mcl-1 in complex with Bim BH3 mutant F4aE | | Descriptor: | ACETATE ION, Bcl-2-like protein 11, Induced myeloid leukemia cell differentiation protein Mcl-1, ... | | Authors: | Fire, E, Grant, R.A, Keating, A.E. | | Deposit date: | 2009-11-02 | | Release date: | 2010-02-16 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (2.351 Å) | | Cite: | Mcl-1-Bim complexes accommodate surprising point mutations via minor structural changes.
Protein Sci., 19, 2010
|
|
4WGI
 
 | | A Single Diastereomer of a Macrolactam Core Binds Specifically to Myeloid Cell Leukemia 1 (MCL1) | | Descriptor: | (2S)-2-[(2S,3R)-10-{[(4-fluorophenyl)sulfonyl]amino}-3-methyl-2-[(methyl{[4-(trifluoromethyl)phenyl]carbamoyl}amino)methyl]-6-oxo-3,4-dihydro-2H-1,5-benzoxazocin-5(6H)-yl]propanoic acid, FORMIC ACID, MAGNESIUM ION, ... | | Authors: | Clifton, M.C, Fairman, J.W, Fang, C, D'Souza, B, Fulroth, B, Leed, A, McCarren, P, Wang, L, Wang, Y, Kaushik, V, Palmer, M, Wei, G, Golub, T.R, Hubbard, B.K, Serrano-Wu, M.H. | | Deposit date: | 2014-09-18 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Single Diastereomer of a Macrolactam Core Binds Specifically to Myeloid Cell Leukemia 1 (MCL1).
Acs Med.Chem.Lett., 5, 2014
|
|
4WMW
 
 | | The structure of MBP-MCL1 bound to ligand 5 at 1.9A | | Descriptor: | 1,2-ETHANEDIOL, 2-hydroxy-5-(methylsulfanyl)benzoic acid, FORMIC ACID, ... | | Authors: | Clifton, M.C, Dranow, D.M. | | Deposit date: | 2014-10-09 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Maltose-Binding Protein Fusion Construct Yields a Robust Crystallography Platform for MCL1.
Plos One, 10, 2015
|
|
4WMS
 
 | | STRUCTURE OF APO MBP-MCL1 AT 1.9A | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, MAGNESIUM ION, ... | | Authors: | Clifton, M.C, Dranow, D.M. | | Deposit date: | 2014-10-09 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Maltose-Binding Protein Fusion Construct Yields a Robust Crystallography Platform for MCL1.
Plos One, 10, 2015
|
|
4WMX
 
 | | The structure of MBP-MCL1 bound to ligand 6 at 2.0A | | Descriptor: | 4-ethenyl-2-[(phenylsulfonyl)amino]benzoic acid, FORMIC ACID, MAGNESIUM ION, ... | | Authors: | Clifton, M.C, Dranow, D.M. | | Deposit date: | 2014-10-09 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Maltose-Binding Protein Fusion Construct Yields a Robust Crystallography Platform for MCL1.
Plos One, 10, 2015
|
|
4WMV
 
 | | STRUCTURE OF MBP-MCL1 BOUND TO ligand 4 AT 2.4A | | Descriptor: | 3-chloro-6-fluoro-1-benzothiophene-2-carboxylic acid, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Clifton, M.C, Moulin, A. | | Deposit date: | 2014-10-09 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A Maltose-Binding Protein Fusion Construct Yields a Robust Crystallography Platform for MCL1.
Plos One, 10, 2015
|
|
4MAN
 
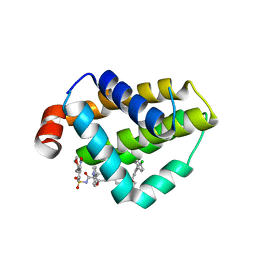 | | Bcl_2-Navitoclax Analog (with Indole) Complex | | Descriptor: | 4-[4-({4'-chloro-3-[2-(dimethylamino)ethoxy]biphenyl-2-yl}methyl)piperazin-1-yl]-2-(1H-indol-5-yloxy)-N-({3-nitro-4-[(tetrahydro-2H-pyran-4-ylmethyl)amino]phenyl}sulfonyl)benzamide, Apoptosis regulator Bcl-2 | | Authors: | Park, C.H. | | Deposit date: | 2013-08-16 | | Release date: | 2014-01-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | ABT-199, a potent and selective BCL-2 inhibitor, achieves antitumor activity while sparing platelets.
NAT.MED. (N.Y.), 19, 2013
|
|
4LVT
 
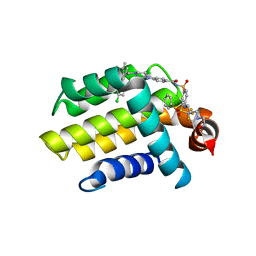 | | Bcl_2-Navitoclax (ABT-263) Complex | | Descriptor: | 4-(4-{[2-(4-chlorophenyl)-5,5-dimethylcyclohex-1-en-1-yl]methyl}piperazin-1-yl)-N-[(4-{[(2R)-4-(morpholin-4-yl)-1-(phenylsulfanyl)butan-2-yl]amino}-3-[(trifluoromethyl)sulfonyl]phenyl)sulfonyl]benzamide, Apoptosis regulator Bcl-2 | | Authors: | Park, C.H. | | Deposit date: | 2013-07-26 | | Release date: | 2013-08-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | ABT-199, a potent and selective BCL-2 inhibitor, achieves antitumor activity while sparing platelets.
NAT.MED. (N.Y.), 19, 2013
|
|
3MQP
 
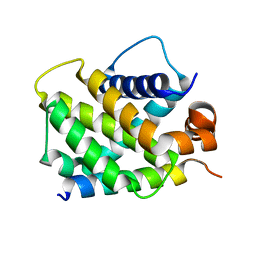 | | Crystal Structure of human BFL-1 in complex with NOXA BH3 peptide, Northeast Structural Genomics Consortium Target HR2930 | | Descriptor: | Bcl-2-related protein A1, Phorbol-12-myristate-13-acetate-induced protein 1 | | Authors: | Guan, R, Xiao, R, Zhao, L, Acton, T.B, Gelinas, C, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-04-28 | | Release date: | 2010-06-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Crystal Structure of human BFL-1 in complex with NOXA BH3 peptide
To be Published
|
|
4PPI
 
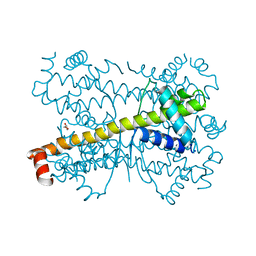 | | Crystal structure of Bcl-xL hexamer | | Descriptor: | Bcl-2-like protein 1, GLYCEROL | | Authors: | Sreekanth, R, Yoon, H.S. | | Deposit date: | 2014-02-27 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.851 Å) | | Cite: | Structural transition in Bcl-xL and its potential association with mitochondrial calcium ion transport
Sci Rep, 5, 2015
|
|
7LH7
 
 | | Crystal structure of BCL-XL in complex with a benzothiazole-based inhibitor | | Descriptor: | Bcl-2-like protein 1, N-(1,3-benzothiazol-2-yl)-2-(4-{[(4-{[(2R)-4-(morpholin-4-yl)-1-(phenylsulfanyl)butan-2-yl]amino}-3-[(trifluoromethyl)sulfonyl]phenyl)sulfonyl]carbamoyl}-1,3-thiazol-2-yl)-1,2,3,4-tetrahydroisoquinoline-8-carboxamide | | Authors: | Judge, R.A, Tao, Z. | | Deposit date: | 2021-01-21 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.409 Å) | | Cite: | Structure-Based Design of A-1293102, a Potent and Selective BCL-XL Inhibitor
ACS Medicinal Chemistry Letters, 12, 2021
|
|
