5TT1
 
 | |
4OVD
 
 | | Crystal structure of a putative peptidoglycan glycosyltransferase from Atopobium parvulum DSM 20469 | | Descriptor: | CALCIUM ION, Peptidoglycan glycosyltransferase | | Authors: | Filippova, E.V, Wawrzak, Z, Kiryukhina, O, Babnigg, G, Clancy, S, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-02-21 | | Release date: | 2014-03-12 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a putative peptidoglycan glycosyltransferase from Atopobium parvulum DSM 20469
To be Published
|
|
7BN9
 
 | |
8TJ3
 
 | |
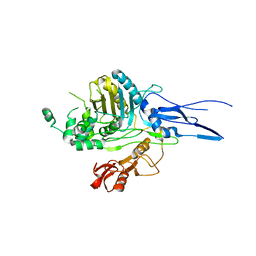
1K25
 
 | |
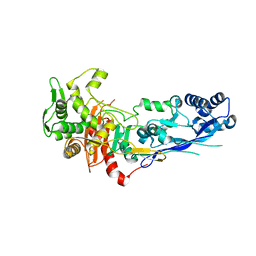
7ZG8
 
 | |
3EQU
 
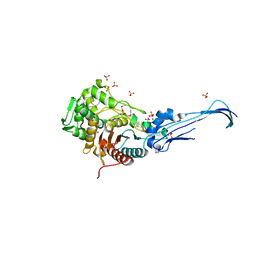 | | Crystal structure of penicillin-binding protein 2 from Neisseria gonorrhoeae | | Descriptor: | GLYCEROL, Penicillin-binding protein 2, SULFATE ION | | Authors: | Powell, A.J, Deacon, A.M, Nicholas, R.A, Davies, C. | | Deposit date: | 2008-10-01 | | Release date: | 2008-10-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of Penicillin-binding Protein 2 from Penicillin-susceptible and -resistant Strains of Neisseria gonorrhoeae Reveal an Unexpectedly Subtle Mechanism for Antibiotic Resistance.
J.Biol.Chem., 284, 2009
|
|
4DKI
 
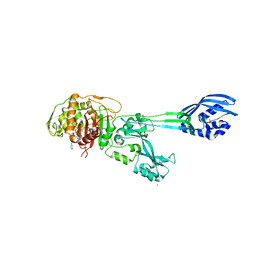 | | Structural Insights into the Anti- Methicillin-Resistant Staphylococcus aureus (MRSA) Activity of Ceftobiprole | | Descriptor: | (2R)-2-[(1R)-1-{[(2Z)-2-(5-amino-1,2,4-thiadiazol-3-yl)-2-(hydroxyimino)acetyl]amino}-2-oxoethyl]-5-({2-oxo-1-[(3R)-pyrrolidin-3-yl]-2,5-dihydro-1H-pyrrol-3-yl}methyl)-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, BICARBONATE ION, CADMIUM ION, ... | | Authors: | Lovering, A.L, Gretes, M.C, Strynadka, N.C.J. | | Deposit date: | 2012-02-03 | | Release date: | 2012-08-01 | | Last modified: | 2012-10-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Insights into the Anti-methicillin-resistant Staphylococcus aureus (MRSA) Activity of Ceftobiprole.
J.Biol.Chem., 287, 2012
|
|
3EQV
 
 | | Crystal structure of penicillin-binding protein 2 from Neisseria gonorrhoeae containing four mutations associated with penicillin resistance | | Descriptor: | GLYCEROL, Penicillin-binding protein 2, SULFATE ION | | Authors: | Powell, A.J, Deacon, A.M, Nicholas, R.A, Davies, C. | | Deposit date: | 2008-10-01 | | Release date: | 2008-10-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of Penicillin-binding Protein 2 from Penicillin-susceptible and -resistant Strains of Neisseria gonorrhoeae Reveal an Unexpectedly Subtle Mechanism for Antibiotic Resistance.
J.Biol.Chem., 284, 2009
|
|
8BH1
 
 | | Core divisome complex FtsWIQBL from Pseudomonas aeruginosa | | Descriptor: | Cell division protein FtsB, Cell division protein FtsL, Cell division protein FtsQ, ... | | Authors: | Kaeshammer, L, van den Ent, F, Jeffery, M, Lowe, J. | | Deposit date: | 2022-10-28 | | Release date: | 2023-04-19 | | Last modified: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structure of the bacterial divisome core complex and antibiotic target FtsWIQBL.
Nat Microbiol, 8, 2023
|
|
4FSF
 
 | | Crystal structure of Pseudomonas aeruginosa PBP3 complexed with compound 14 | | Descriptor: | (4R,5S,8Z)-8-(2-amino-1,3-thiazol-4-yl)-1-[3-(1,5-dihydroxy-4-oxo-1,4-dihydropyridin-2-yl)-1,2-oxazol-5-yl]-5-formyl-11,11-dimethyl-1,7-dioxo-4-(sulfoamino)-10-oxa-2,6,9-triazadodec-8-en-12-oic acid, Penicillin-binding protein 3 | | Authors: | Han, S. | | Deposit date: | 2012-06-27 | | Release date: | 2012-10-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Novel monobactams utilizing a siderophore uptake mechanism for the treatment of gram-negative infections.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
2WAD
 
 | | PENICILLIN-BINDING PROTEIN 2B (PBP-2B) FROM STREPTOCOCCUS PNEUMONIAE (STRAIN 5204) | | Descriptor: | PENICILLIN-BINDING PROTEIN 2B, ZINC ION | | Authors: | Contreras-Martel, C, Dahout-Gonzalez, C, Dos-Santos-Martins, A, Kotnik, M, Dessen, A. | | Deposit date: | 2009-02-05 | | Release date: | 2009-02-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Pbp Active Site Flexibility as the Key Mechanism for Beta-Lactam Resistance in Pneumococci
J.Mol.Biol., 387, 2009
|
|
2WAF
 
 | | PENICILLIN-BINDING PROTEIN 2B (PBP-2B) FROM STREPTOCOCCUS PNEUMONIAE (STRAIN R6) | | Descriptor: | CHLORIDE ION, PENICILLIN-BINDING PROTEIN 2B | | Authors: | Contreras-Martel, C, Dahout-Gonzalez, C, Dos-Santos-Martins, A, Kotnik, M, Dessen, A. | | Deposit date: | 2009-02-05 | | Release date: | 2009-02-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Pbp Active Site Flexibility as the Key Mechanism for Beta-Lactam Resistance in Pneumococci
J.Mol.Biol., 387, 2009
|
|
2WAE
 
 | | PENICILLIN-BINDING PROTEIN 2B (PBP-2B) FROM STREPTOCOCCUS PNEUMONIAE (STRAIN 5204) | | Descriptor: | PENICILLIN-BINDING PROTEIN 2B, ZINC ION | | Authors: | Contreras-Martel, C, Dahout-Gonzalez, C, Dos-Santos-Martins, A, Kotnik, M, Dessen, A. | | Deposit date: | 2009-02-05 | | Release date: | 2009-02-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Pbp Active Site Flexibility as the Key Mechanism for Beta-Lactam Resistance in Pneumococci
J.Mol.Biol., 387, 2009
|
|
3OC2
 
 | | Crystal structure of penicillin-binding protein 3 from Pseudomonas aeruginosa | | Descriptor: | CHLORIDE ION, Penicillin-binding protein 3 | | Authors: | Sainsbury, S, Bird, L, Stuart, D.I, Owens, R.J, Ren, J, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2010-08-09 | | Release date: | 2010-11-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.968 Å) | | Cite: | Crystal structures of penicillin-binding protein 3 from Pseudomonas aeruginosa: comparison of native and antibiotic-bound forms
J.Mol.Biol., 405, 2011
|
|
3OCN
 
 | | Crystal structure of penicillin-binding protein 3 from Pseudomonas aeruginosa in complex with ceftazidime | | Descriptor: | 1-({(2R)-2-[(1R)-1-{[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-{[(2-carboxypropan-2-yl)oxy]imino}acetyl]amino}-2-oxoethyl]-4-carboxy-3,6-dihydro-2H-1,3-thiazin-5-yl}methyl)pyridinium, penicillin-binding protein 3 | | Authors: | Sainsbury, S, Bird, L, Stuart, D.I, Owens, R.J, Ren, J, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2010-08-10 | | Release date: | 2010-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal structures of penicillin-binding protein 3 from Pseudomonas aeruginosa: comparison of native and antibiotic-bound forms
J.Mol.Biol., 405, 2011
|
|
3OCL
 
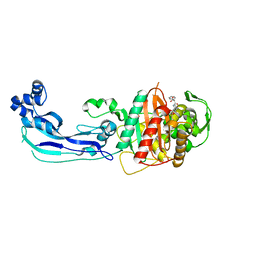 | | Crystal structure of penicillin-binding protein 3 from Pseudomonas aeruginosa in complex with carbenicillin | | Descriptor: | (2R,4S)-2-[(1R)-1-{[(2S)-2-carboxy-2-phenylacetyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Sainsbury, S, Bird, L, Stuart, D.I, Owens, R.J, Ren, J, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2010-08-10 | | Release date: | 2010-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of penicillin-binding protein 3 from Pseudomonas aeruginosa: comparison of native and antibiotic-bound forms
J.Mol.Biol., 405, 2011
|
|
6BSR
 
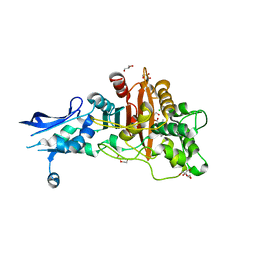 | | Crystal structure of penicillin-binding protein 4 (PBP4) from Enterococcus faecalis in the benzylpenicillin bound form. | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Moon, T.M, D'Andrea, E.D, Peti, W, Page, R. | | Deposit date: | 2017-12-04 | | Release date: | 2018-10-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | The structures of penicillin-binding protein 4 (PBP4) and PBP5 fromEnterococciprovide structural insights into beta-lactam resistance.
J. Biol. Chem., 293, 2018
|
|
6C84
 
 | |
6BSQ
 
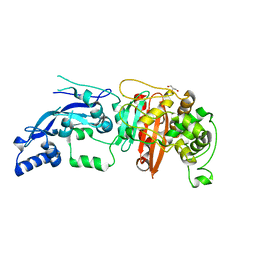 | | Enterococcus faecalis Penicillin Binding Protein 4 (PBP4) | | Descriptor: | CHLORIDE ION, GLYCEROL, PBP4 protein | | Authors: | Moon, T.M, D'Andrea, E.D, Peti, W, Page, R. | | Deposit date: | 2017-12-04 | | Release date: | 2018-10-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structures of penicillin-binding protein 4 (PBP4) and PBP5 fromEnterococciprovide structural insights into beta-lactam resistance.
J. Biol. Chem., 293, 2018
|
|
4WEJ
 
 | | Crystal structure of Pseudomonas aeruginosa PBP3 with a R4 substituted allyl monocarbam | | Descriptor: | (3R,4S,7Z)-7-(2-amino-1,3-thiazol-4-yl)-4-formyl-1-[({3-[(5R)-5-hydroxy-4-oxo-4,5-dihydropyridin-2-yl]-4-[3-(methylsulfonyl)propyl]-5-oxo-4,5-dihydro-1H-1,2,4-triazol-1-yl}sulfonyl)amino]-10,10-dimethyl-1,6-dioxo-3-(prop-2-en-1-yl)-9-oxa-2,5,8-triazaundec-7-en-11-oic acid, Penicillin-binding protein 3 | | Authors: | Ferguson, A.D. | | Deposit date: | 2014-09-10 | | Release date: | 2015-04-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.045 Å) | | Cite: | SAR and Structural Analysis of Siderophore-Conjugated Monocarbam Inhibitors of Pseudomonas aeruginosa PBP3.
Acs Med.Chem.Lett., 6, 2015
|
|
4WEL
 
 | | Crystal structure of Pseudomonas aeruginosa PBP3 with SMC-3176 | | Descriptor: | (3S,4S,7Z)-7-(2-amino-1,3-thiazol-4-yl)-4-formyl-1-[({3-(5-hydroxy-4-oxo-3,4-dihydropyridin-2-yl)-4-[3-(methylsulfonyl)propyl]-5-oxo-4,5-dihydro-1H-1,2,4-triazol-1-yl}sulfonyl)amino]-3,10,10-trimethyl-1,6-dioxo-9-oxa-2,5,8-triazaundec-7-en-11-oic acid, Penicillin-binding protein 3 | | Authors: | Ferguson, A.D. | | Deposit date: | 2014-09-10 | | Release date: | 2015-04-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | SAR and Structural Analysis of Siderophore-Conjugated Monocarbam Inhibitors of Pseudomonas aeruginosa PBP3.
Acs Med.Chem.Lett., 6, 2015
|
|
4WEK
 
 | | Crystal structure of Pseudomonas aeruginosa PBP3 with a R4 substituted vinyl monocarbam | | Descriptor: | (3S,4S,7Z)-7-(2-amino-1,3-thiazol-4-yl)-3-ethenyl-4-formyl-1-[({3-(5-hydroxy-4-oxo-3,4-dihydropyridin-2-yl)-4-[3-(methylsulfonyl)propyl]-5-oxo-4,5-dihydro-1H-1,2,4-triazol-1-yl}sulfonyl)amino]-10,10-dimethyl-1,6-dioxo-9-oxa-2,5,8-triazaundec-7-en-11-oic acid, Penicillin-binding protein 3 | | Authors: | Ferguson, A.D. | | Deposit date: | 2014-09-10 | | Release date: | 2015-04-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | SAR and Structural Analysis of Siderophore-Conjugated Monocarbam Inhibitors of Pseudomonas aeruginosa PBP3.
Acs Med.Chem.Lett., 6, 2015
|
|
1QME
 
 | | PENICILLIN-BINDING PROTEIN 2X (PBP-2X) | | Descriptor: | PENICILLIN-BINDING PROTEIN 2X, SULFATE ION | | Authors: | Gordon, E.J, Mouz, N, Duee, E, Dideberg, O. | | Deposit date: | 1999-09-28 | | Release date: | 2000-05-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Crystal Structure of the Penicillin-Binding Protein 2X from Streptococcus Pneumoniae and its Acyl-Enzyme Form: Implication in Drug Resistance.
J.Mol.Biol., 299, 2000
|
|
1QMF
 
 | | PENICILLIN-BINDING PROTEIN 2X (PBP-2X) ACYL-ENZYME COMPLEX | | Descriptor: | 2-[CARBOXY-(2-FURAN-2-YL-2-METHOXYIMINO-ACETYLAMINO)-METHYL]-5-METHYL-3,6-DIHYDRO-2H-[1,3]THIAZINE-4-CARBOXYLIC ACID, CEFUROXIME (OCT-3-ENE FORM), PENICILLIN-BINDING PROTEIN 2X | | Authors: | Gordon, E.J, Mouz, N, Duee, E, Dideberg, O. | | Deposit date: | 1999-09-28 | | Release date: | 2000-05-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Crystal Structure of the Penicillin Binding Protein 2X from Streptococcus Pneumoniae and its Acyl-Enzyme Form: Implication in Drug Resistance
J.Mol.Biol., 299, 2000
|
|
