2A5D
 
 | | Structural basis for the activation of cholera toxin by human ARF6-GTP | | Descriptor: | ADP-ribosylation factor 6, Cholera enterotoxin, A chain, ... | | Authors: | O'Neal, C.J, Jobling, M.G, Holmes, R.K, Hol, W.G.J. | | Deposit date: | 2005-06-30 | | Release date: | 2005-08-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the activation of cholera toxin by human ARF6-GTP.
Science, 309, 2005
|
|
3N5C
 
 | | Crystal Structure of Arf6DELTA13 complexed with GDP | | Descriptor: | ADP-ribosylation factor 6, CHLORIDE ION, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Aizel, K, Biou, V, Cherfils, J. | | Deposit date: | 2010-05-25 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | SAXS and X-ray crystallography suggest an unfolding model for the GDP/GTP conformational switch of the small GTPase Arf6.
J.Mol.Biol., 402, 2010
|
|
4KAX
 
 | | Crystal structure of the Grp1 PH domain in complex with Arf6-GTP | | Descriptor: | ADP-ribosylation factor 6, CITRIC ACID, Cytohesin-3, ... | | Authors: | Lambright, D.G, Malaby, A.W, van den Berg, B. | | Deposit date: | 2013-04-23 | | Release date: | 2013-08-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for membrane recruitment and allosteric activation of cytohesin family Arf GTPase exchange factors.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6PAU
 
 | |
2W83
 
 | | Crystal structure of the ARF6 GTPase in complex with a specific effector, JIP4 | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, ADP-RIBOSYLATION FACTOR 6, C-JUN-AMINO-TERMINAL KINASE-INTERACTING PROTEIN 4, ... | | Authors: | Isabet, T, Montagnac, G, Regazzoni, K, Raynal, B, El Khadali, F, Franco, M, England, P, Chavrier, P, Houdusse, A, Menetrey, J. | | Deposit date: | 2009-01-08 | | Release date: | 2009-07-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | The Structural Basis of Arf Effector Specificity: The Crystal Structure of Arf6 in a Complex with Jip4.
Embo J., 28, 2009
|
|
2A5F
 
 | | Cholera toxin A1 subunit bound to its substrate, NAD+, and its human protein activator, ARF6 | | Descriptor: | ADP-ribosylation factor 6, Cholera enterotoxin, A chain, ... | | Authors: | O'Neal, C.J, Jobling, M.G, Holmes, R.K, Hol, W.G.J. | | Deposit date: | 2005-06-30 | | Release date: | 2005-08-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural basis for the activation of cholera toxin by human ARF6-GTP.
Science, 309, 2005
|
|
7RK3
 
 | |
2B0O
 
 | | Crystal structure of UPLC1 GAP domain | | Descriptor: | UPLC1, ZINC ION | | Authors: | Ismail, S, Shen, L, Arrowsmith, C, Edwards, A, Sundstrom, M, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-09-14 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural analysis of GAP and ankyrin domains of UPLC1
To be Published
|
|
1E0S
 
 | | small G protein Arf6-GDP | | Descriptor: | ADP-ribosylation factor 6, AMMONIUM ION, BETA-MERCAPTOETHANOL, ... | | Authors: | Menetrey, J, Cherfils, J. | | Deposit date: | 2000-04-06 | | Release date: | 2000-04-18 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structure of Arf6-Gdp Suggests a Basis for Guanine Nucleotide Exchange Factors Specificity
Nat.Struct.Biol., 7, 2000
|
|
3PCR
 
 | | Structure of EspG-Arf6 complex | | Descriptor: | ADP-ribosylation factor 6, EspG, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Tomchick, D.R, Alto, N.M, Selyunin, A.S. | | Deposit date: | 2010-10-21 | | Release date: | 2011-01-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The assembly of a GTPase-kinase signalling complex by a bacterial catalytic scaffold.
Nature, 469, 2011
|
|
6PAV
 
 | |
2A5G
 
 | | Cholera toxin A1 subunit bound to ARF6(Q67L) | | Descriptor: | ADP-ribosylation factor 6, Cholera enterotoxin, A chain, ... | | Authors: | O'Neal, C.J, Jobling, M.G, Holmes, R.K, Hol, W.G.J. | | Deposit date: | 2005-06-30 | | Release date: | 2005-08-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Structural basis for the activation of cholera toxin by human ARF6-GTP.
Science, 309, 2005
|
|
2J5X
 
 | | STRUCTURE OF THE SMALL G PROTEIN ARF6 IN COMPLEX WITH GTPGAMMAS | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, ADP-RIBOSYLATION FACTOR 6, MAGNESIUM ION | | Authors: | Pasqualato, S, Menetrey, J, Franco, M, Cherfils, J. | | Deposit date: | 2006-09-20 | | Release date: | 2006-09-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Structural Gdp-GTP Cycle of Human Arf6.
Embo Rep., 2, 2001
|
|
3LVQ
 
 | | The crystal structure of ASAP3 in complex with Arf6 in transition state | | Descriptor: | ALUMINUM FLUORIDE, Arf-GAP with SH3 domain, ANK repeat and PH domain-containing protein 3, ... | | Authors: | Ismail, S.A, Vetter, I.R, Sot, B, Wittinghofer, A. | | Deposit date: | 2010-02-22 | | Release date: | 2010-06-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.38 Å) | | Cite: | The structure of an Arf-ArfGAP complex reveals a Ca2+ regulatory mechanism
Cell(Cambridge,Mass.), 141, 2010
|
|
3LVR
 
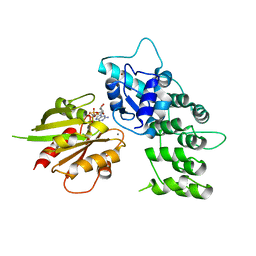 | | The crystal structure of ASAP3 in complex with Arf6 in transition state soaked with Calcium | | Descriptor: | ALUMINUM FLUORIDE, Arf-GAP with SH3 domain, ANK repeat and PH domain-containing protein 3, ... | | Authors: | Ismail, S.A, Vetter, I.R, Sot, B, Wittinghofer, A. | | Deposit date: | 2010-02-22 | | Release date: | 2010-06-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.38 Å) | | Cite: | The structure of an Arf-ArfGAP complex reveals a Ca2+ regulatory mechanism
Cell(Cambridge,Mass.), 141, 2010
|
|
7XRD
 
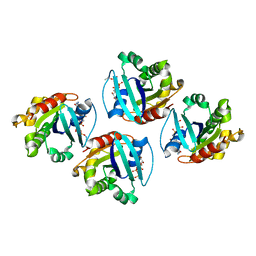 | |
4FME
 
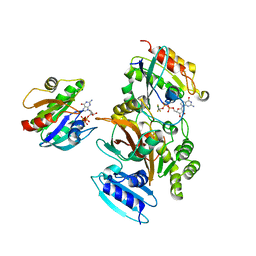 | | EspG-Rab1-Arf6 complex | | Descriptor: | ADP-ribosylation factor 6, ALUMINUM FLUORIDE, EspG protein, ... | | Authors: | Shao, F, Zhu, Y. | | Deposit date: | 2012-06-16 | | Release date: | 2012-09-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Structurally Distinct Bacterial TBC-like GAPs Link Arf GTPase to Rab1 Inactivation to Counteract Host Defenses.
Cell(Cambridge,Mass.), 150, 2012
|
|
6BBP
 
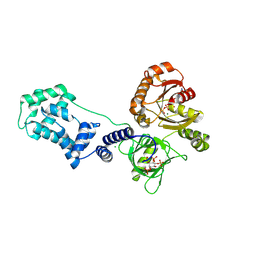 | | Model for compact volume of truncated monomeric Cytohesin-3 (Grp1; amino acids 63-399) E161A 6GS Arf6 Q67L fusion protein | | Descriptor: | Cytohesin-3,ADP-ribosylation factor 6, GUANOSINE-5'-TRIPHOSPHATE, INOSITOL-(1,3,4,5)-TETRAKISPHOSPHATE, ... | | Authors: | Das, S, Malaby, A.W, Lambright, D.G. | | Deposit date: | 2017-10-19 | | Release date: | 2018-01-10 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (35 Å) | | Cite: | Structural Dynamics Control Allosteric Activation of Cytohesin Family Arf GTPase Exchange Factors.
Structure, 26, 2018
|
|
6BBQ
 
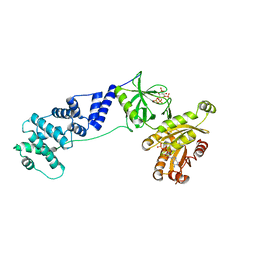 | | Model for extended volume of truncated monomeric Cytohesin-3 (Grp1; amino acids 63-399) E161A Arf6 Q67L fusion protein | | Descriptor: | Cytohesin-3,ADP-ribosylation factor 6, GUANOSINE-5'-TRIPHOSPHATE, INOSITOL-(1,3,4,5)-TETRAKISPHOSPHATE, ... | | Authors: | Das, S, Malaby, A.W, Lambright, D.G. | | Deposit date: | 2017-10-19 | | Release date: | 2018-01-10 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (35 Å) | | Cite: | Structural Dynamics Control Allosteric Activation of Cytohesin Family Arf GTPase Exchange Factors.
Structure, 26, 2018
|
|
2BAO
 
 | |
2BAU
 
 | |
