2X4W
 
 | | Molecular basis of Histone H3K36me3 recognition by the PWWP domain of BRPF1. | | Descriptor: | FORMIC ACID, HISTONE H3.2, PEREGRIN | | Authors: | Vezzoli, A, Bonadies, N, Allen, M.D, Freund, S.M.V, Santiveri, C.M, Kvinlaug, B, Huntly, B.J.P, Gottgens, B, Bycroft, M. | | Deposit date: | 2010-02-02 | | Release date: | 2010-04-21 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular Basis of Histone H3K36Me3 Recognition by the Pwwp Domain of Brpf1.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3MO8
 
 | | PWWP Domain of Human Bromodomain and PHD finger-containing protein 1 In Complex with Trimethylated H3K36 Peptide | | Descriptor: | Histone H3.2 TRIMETHYLATED H3K36 PEPTIDE, Peregrin | | Authors: | Lam, R, Zeng, H, Ni, S, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-04-22 | | Release date: | 2010-06-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural and Histone Binding Ability Characterizations of Human PWWP Domains.
Plos One, 6, 2011
|
|
4MZG
 
 | | Crystal structure of human Spindlin1 bound to histone H3K4me3 peptide | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, ... | | Authors: | Su, X, Ding, X, Li, H. | | Deposit date: | 2013-09-30 | | Release date: | 2014-03-26 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Molecular basis underlying histone H3 lysine-arginine methylation pattern readout by Spin/Ssty repeats of Spindlin1
Genes Dev., 28, 2014
|
|
2X4Y
 
 | | Molecular basis of Histone H3K36me3 recognition by the PWWP domain of BRPF1. | | Descriptor: | HISTONE H3.2, PEREGRIN, SULFATE ION | | Authors: | Vezzoli, A, Bonadies, N, Allen, M.D, Freund, S.M.V, Santiveri, C.M, Kvinlaug, B, Huntly, B.J.P, Gottgens, B, Bycroft, M. | | Deposit date: | 2010-02-02 | | Release date: | 2010-04-21 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular Basis of Histone H3K36Me3 Recognition by the Pwwp Domain of Brpf1.
Nat.Struct.Mol.Biol., 17, 2010
|
|
6WAU
 
 | | Complex structure of PHF19 | | Descriptor: | Histone H3.1t peptide, PHD finger protein 19, UNKNOWN ATOM OR ION | | Authors: | Dong, C, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Min, J.R, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-03-26 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for histone variant H3tK27me3 recognition by PHF1 and PHF19.
Elife, 9, 2020
|
|
6WAT
 
 | | complex structure of PHF1 | | Descriptor: | Histone H3.1t peptide, PHD finger protein 1, UNKNOWN ATOM OR ION | | Authors: | Dong, C, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Min, J.R, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-03-26 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for histone variant H3tK27me3 recognition by PHF1 and PHF19.
Elife, 9, 2020
|
|
4V2W
 
 | | JMJD2A COMPLEXED WITH NI(II), NOG AND HISTONE H3K27me3 PEPTIDE (16-35) | | Descriptor: | CHLORIDE ION, GLYCEROL, HISTONE H3.1T, ... | | Authors: | Chowdhury, R, Zafred, D, Schofield, C.J. | | Deposit date: | 2014-10-15 | | Release date: | 2014-11-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Studies on the Catalytic Domains of Multiple Jmjc Oxygenases Using Peptide Substrates.
Epigenetics, 9, 2014
|
|
2X4X
 
 | | Molecular basis of Histone H3K36me3 recognition by the PWWP domain of BRPF1. | | Descriptor: | HISTONE H3.2, PEREGRIN, SULFATE ION | | Authors: | Vezzoli, A, Bonadies, N, Allen, M.D, Freund, S.M.V, Santiveri, C.M, Kvinlaug, B, Huntly, B.J.P, Gottgens, B, Bycroft, M. | | Deposit date: | 2010-02-02 | | Release date: | 2010-04-21 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular Basis of Histone H3K36Me3 Recognition by the Pwwp Domain of Brpf1.
Nat.Struct.Mol.Biol., 17, 2010
|
|
4OUC
 
 | | Structure of human haspin in complex with histone H3 substrate | | Descriptor: | (2R,3R,4S,5R)-2-(4-AMINO-5-IODO-7H-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-(HYDROXYMETHYL)TETRAHYDROFURAN-3,4-DIOL, 1,2-ETHANEDIOL, Histone H3.2, ... | | Authors: | Chaikuad, A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-02-15 | | Release date: | 2014-04-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Modulation of the chromatin phosphoproteome by the haspin protein kinase.
Mol Cell Proteomics, 13, 2014
|
|
4A7J
 
 | | Symmetric Dimethylation of H3 Arginine 2 is a Novel Histone Mark that Supports Euchromatin Maintenance | | Descriptor: | HISTONE H3.1T, WD REPEAT-CONTAINING PROTEIN 5 | | Authors: | Migliori, V, Muller, J, Phalke, S, Low, D, Bezzi, M, ChuenMok, W, Gunaratne, J, Capasso, P, Bassi, C, Cecatiello, V, DeMarco, A, Blackstock, W, Kuznetsov, V, Amati, B, Mapelli, M, Guccione, E. | | Deposit date: | 2011-11-14 | | Release date: | 2012-01-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Symmetric Dimethylation of H3R2 is a Newly Identified Histone Mark that Supports Euchromatin Maintenance
Nat.Struct.Mol.Biol., 19, 2012
|
|
5VAC
 
 | | Crystal Structure of ATXR5 SET domain in complex with K36me3 histone H3 peptide | | Descriptor: | DIMETHYL SULFOXIDE, Histone H3.2, Probable Histone-lysine N-methyltransferase ATXR5, ... | | Authors: | Bergamin, E, Sarvan, S, Malette, J, Eram, M, Yeung, S, Mongeon, V, Joshi, M, Brunzelle, J.S, Michaels, S.D, Blais, A, Vedadi, M, Couture, J.F. | | Deposit date: | 2017-03-24 | | Release date: | 2017-04-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.949 Å) | | Cite: | Molecular basis for the methylation specificity of ATXR5 for histone H3.
Nucleic Acids Res., 45, 2017
|
|
3T6R
 
 | | Structure of UHRF1 in complex with unmodified H3 N-terminal tail | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, Histone H3.1t N-terminal peptide, MAGNESIUM ION, ... | | Authors: | Xie, S, Jakoncic, J, Qian, C.M. | | Deposit date: | 2011-07-29 | | Release date: | 2011-11-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | UHRF1 double tudor domain and the adjacent PHD finger act together to recognize K9me3-containing histone H3 tail
J.Mol.Biol., 415, 2012
|
|
7UVA
 
 | | Crystal structure of KDM2A histone demethylase catalytic domain in complex with an H3C36 peptide modified by UNC8015 | | Descriptor: | FE (III) ION, Histone H3.2, Lysine-specific demethylase 2A, ... | | Authors: | Budziszewski, G.R, Azzam, D.N, Spangler, C.J, Skrajna, A, Foley, C.A, James, L.I, Frye, S.V, McGinty, R.K. | | Deposit date: | 2022-04-29 | | Release date: | 2023-02-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural basis of paralog-specific KDM2A/B nucleosome recognition.
Nat.Chem.Biol., 19, 2023
|
|
6ACE
 
 | |
5B0Z
 
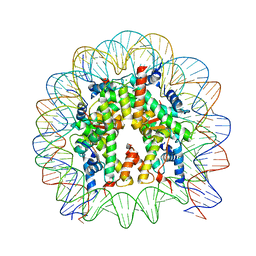 | | The crystal structure of the nucleosome containing H3.2, at 1.98 A resolution | | Descriptor: | CHLORIDE ION, DNA (146-MER), Histone H2A type 1-B/E, ... | | Authors: | Suzuki, Y, Horikoshi, N, Kato, D, Kurumizaka, H. | | Deposit date: | 2015-11-14 | | Release date: | 2016-01-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.987 Å) | | Cite: | Crystal structure of the nucleosome containing histone H3 with crotonylated lysine 122
Biochem.Biophys.Res.Commun., 469, 2016
|
|
4V2V
 
 | | JMJD2A COMPLEXED WITH NI(II), NOG AND HISTONE H3K27me3 PEPTIDE (25-29) ARK(me3)SA | | Descriptor: | CHLORIDE ION, HISTONE H3.1T, LYSINE-SPECIFIC DEMETHYLASE 4A, ... | | Authors: | Chowdhury, R, Madden, S.K, Schofield, C.J. | | Deposit date: | 2014-10-15 | | Release date: | 2014-11-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Studies on the Catalytic Domains of Multiple Jmjc Oxygenases Using Peptide Substrates.
Epigenetics, 9, 2014
|
|
2YBP
 
 | | JMJD2A COMPLEXED WITH R-2-HYDROXYGLUTARATE AND HISTONE H3K36me3 PEPTIDE (30-41) | | Descriptor: | (2R)-2-hydroxypentanedioic acid, GLYCEROL, HISTONE H3.1T, ... | | Authors: | Chowdhury, R, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2011-03-09 | | Release date: | 2011-03-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases.
EMBO Rep., 12, 2011
|
|
3R93
 
 | | Crystal structure of the chromo domain of M-phase phosphoprotein 8 bound to H3K9Me3 peptide | | Descriptor: | H3K9Me3 peptide, M-phase phosphoprotein 8, UNKNOWN ATOM OR ION | | Authors: | Li, J, Li, Z, Ruan, J, Xu, C, Tong, Y, Pan, P.W, Tempel, W, Crombet, L, Min, J, Zang, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-03-24 | | Release date: | 2011-04-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.057 Å) | | Cite: | Structural basis for specific binding of human MPP8 chromodomain to histone H3 methylated at lysine 9.
Plos One, 6, 2011
|
|
6OIE
 
 | |
4MZF
 
 | | Crystal structure of human Spindlin1 bound to histone H3(K4me3-R8me2a) peptide | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Peptide from Histone H3.2, ... | | Authors: | Su, X, Ding, X, Li, H. | | Deposit date: | 2013-09-30 | | Release date: | 2014-03-26 | | Method: | X-RAY DIFFRACTION (2.098 Å) | | Cite: | Molecular basis underlying histone H3 lysine-arginine methylation pattern readout by Spin/Ssty repeats of Spindlin1
Genes Dev., 28, 2014
|
|
4MZH
 
 | |
5CIU
 
 | | Structural basis of the recognition of H3K36me3 by DNMT3B PWWP domain | | Descriptor: | DNA (cytosine-5)-methyltransferase 3B, GLYCEROL, Histone H3.2 | | Authors: | Rondelet, G, DAL MASO, T, Willems, L, Wouters, J. | | Deposit date: | 2015-07-13 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structural basis for recognition of histone H3K36me3 nucleosome by human de novo DNA methyltransferases 3A and 3B.
J.Struct.Biol., 194, 2016
|
|
2YBS
 
 | |
8JLB
 
 | | Cryo-EM structure of the 145 bp human nucleosome containing H3.2 C110A mutant | | Descriptor: | DNA (145-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Oishi, T, Hatazawa, S, Kujirai, T, Kato, J, Kobayashi, Y, Ogasawara, M, Akatsu, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-06-02 | | Release date: | 2023-10-04 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.36 Å) | | Cite: | Contributions of histone tail clipping and acetylation in nucleosome transcription by RNA polymerase II.
Nucleic Acids Res., 51, 2023
|
|
3DB3
 
 | | Crystal structure of the tandem tudor domains of the E3 ubiquitin-protein ligase UHRF1 in complex with trimethylated histone H3-K9 peptide | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, Trimethylated histone H3-K9 peptide | | Authors: | Walker, J.R, Avvakumov, G.V, Xue, S, Dong, A, Li, Y, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-05-30 | | Release date: | 2008-09-16 | | Last modified: | 2012-04-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Recognition of multivalent histone states associated with heterochromatin by UHRF1 protein.
J.Biol.Chem., 286, 2011
|
|
