6IBE
 
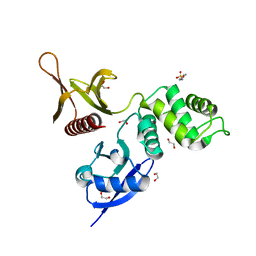 | | The FERM domain of Human EPB41L3 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Band 4.1-like protein 3 | | Authors: | Bradshaw, W.J, Katis, V.L, Newman, J.A, Fernandez-Cid, A, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2018-11-29 | | Release date: | 2018-12-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The FERM domain of Human EPB41L3
To Be Published
|
|
4YL8
 
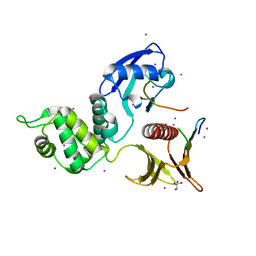 | | Crystal structure of the Crumbs/Moesin complex | | Descriptor: | GLYCEROL, IODIDE ION, Moesin, ... | | Authors: | Wei, Z, Li, Y, Zhang, M. | | Deposit date: | 2015-03-05 | | Release date: | 2015-04-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis for the Phosphorylation-regulated Interaction between the Cytoplasmic Tail of Cell Polarity Protein Crumbs and the Actin-binding Protein Moesin
J.Biol.Chem., 290, 2015
|
|
7LWH
 
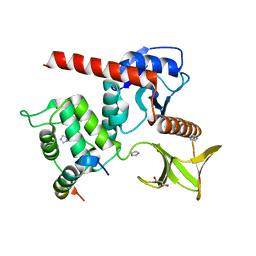 | | Human neurofibromin 2/merlin residues 1-339 in complex with LATS1 | | Descriptor: | GLYCEROL, IMIDAZOLE, Merlin, ... | | Authors: | Primi, M.C, Rangarajan, E.S, Izard, T. | | Deposit date: | 2021-03-01 | | Release date: | 2021-08-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.606 Å) | | Cite: | Conformational flexibility determines the Nf2/merlin tumor suppressor functions.
Matrix Biol Plus, 12, 2021
|
|
1SGH
 
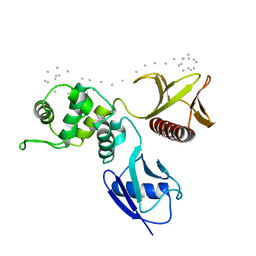 | | Moesin FERM domain bound to EBP50 C-terminal peptide | | Descriptor: | Ezrin-radixin-moesin binding phosphoprotein 50, Moesin | | Authors: | Finnerty, C.M, Chambers, D, Ingraffea, J, Faber, H.R, Karplus, P.A, Bretscher, A. | | Deposit date: | 2004-02-23 | | Release date: | 2004-06-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The EBP50-moesin interaction involves a binding site regulated by direct masking on the FERM domain
J.Cell.Sci., 117, 2004
|
|
6QLY
 
 | | IDOL FERM domain | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase MYLIP, SULFATE ION | | Authors: | Martinelli, L, Sixma, T.K. | | Deposit date: | 2019-02-01 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural analysis of the LDL receptor-interacting FERM domain in the E3 ubiquitin ligase IDOL reveals an obscured substrate-binding site.
J.Biol.Chem., 295, 2020
|
|
7EDR
 
 | |
3BIN
 
 | | Structure of the DAL-1 and TSLC1 (372-383) complex | | Descriptor: | Band 4.1-like protein 3, Cell adhesion molecule 1 | | Authors: | Busam, R.D, Arrowsmith, C.H, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Kallas, A, Karlberg, T, Kotenyova, T, Lehtio, L, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Sagemark, J, Svensson, L, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Welin, M, Berglund, H, Persson, C, Hallberg, B.M. | | Deposit date: | 2007-11-30 | | Release date: | 2008-01-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of tumor suppressor in lung cancer 1 (TSLC1) binding to differentially expressed in adenocarcinoma of the lung (DAL-1/4.1B)
J.Biol.Chem., 286, 2011
|
|
8Y8Y
 
 | | Crystal structure of the PTPN21 FERM domain | | Descriptor: | CHLORIDE ION, Tyrosine-protein phosphatase non-receptor type 21 | | Authors: | Lee, H.S, Ku, B. | | Deposit date: | 2024-02-06 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Structural analysis of the FERM domain of human protein tyrosine phosphatase non-receptor type 21.
Acta Crystallogr.,Sect.F, 80, 2024
|
|
4P7I
 
 | | Crystal structure of the Merlin FERM/DCAF1 complex | | Descriptor: | GLYCEROL, Merlin, Protein VPRBP | | Authors: | Wei, Z, Li, Y, Zhang, M. | | Deposit date: | 2014-03-27 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of the binding of Merlin FERM domain to the E3 ubiquitin ligase substrate adaptor DCAF1.
J.Biol.Chem., 289, 2014
|
|
3QIJ
 
 | | Primitive-monoclinic crystal structure of the FERM domain of protein 4.1R | | Descriptor: | Protein 4.1, UNKNOWN ATOM OR ION | | Authors: | Nedyalkova, L, Zhong, N, Tong, Y, Tempel, W, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-01-27 | | Release date: | 2011-02-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Primitive-monoclinic crystal structure of the FERM domain of protein 4.1R
to be published
|
|
1NI2
 
 | | Structure of the active FERM domain of Ezrin | | Descriptor: | Ezrin | | Authors: | Smith, W.J, Nassar, N, Bretscher, A.P, Cerione, R.A, Karplus, P.A. | | Deposit date: | 2002-12-20 | | Release date: | 2003-02-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the Active N-terminal Domain of Ezrin. CONFORMATIONAL AND MOBILITY CHANGES IDENTIFY KEYSTONE INTERACTIONS.
J.Biol.Chem., 278, 2003
|
|
2YVC
 
 | |
1ISN
 
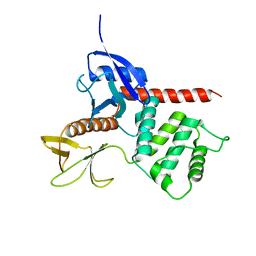 | | Crystal structure of merlin FERM domain | | Descriptor: | merlin | | Authors: | Shimizu, T, Seto, A, Maita, N, Hamada, K, Tsukita, S, Tsukita, S, Hakoshima, T. | | Deposit date: | 2001-12-13 | | Release date: | 2002-04-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for neurofibromatosis type 2. Crystal structure of the merlin FERM domain.
J.Biol.Chem., 277, 2002
|
|
2ZPY
 
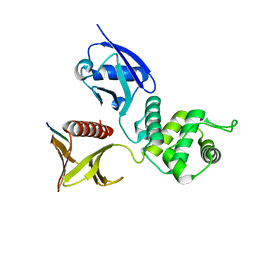 | | Crystal structure of the mouse radxin FERM domain complexed with the mouse CD44 cytoplasmic peptide | | Descriptor: | CD44 antigen, Radixin | | Authors: | Mori, T, Kitano, K, Terawaki, S, Maesaki, R, Fukami, Y, Hakoshima, T. | | Deposit date: | 2008-07-31 | | Release date: | 2008-08-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for CD44 recognition by ERM proteins
J.Biol.Chem., 283, 2008
|
|
1J19
 
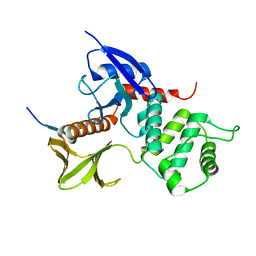 | | Crystal structure of the radxin FERM domain complexed with the ICAM-2 cytoplasmic peptide | | Descriptor: | 16-mer peptide from Intercellular adhesion molecule-2, radixin | | Authors: | Hamada, K, Shimizu, T, Yonemura, S, Tsukita, S, Tsukita, S, Hakoshima, T. | | Deposit date: | 2002-12-02 | | Release date: | 2003-03-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of adhesion-molecule recognition by ERM proteins revealed by the crystal structure of the radixin-ICAM-2 complex
EMBO J., 22, 2003
|
|
8GVL
 
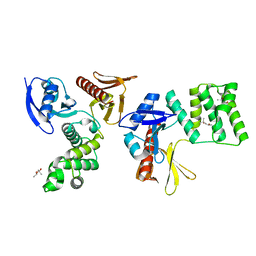 | | PTPN21 FERM | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Tyrosine-protein phosphatase non-receptor type 21 | | Authors: | Chen, L, Zheng, Y.Y, Zhou, C. | | Deposit date: | 2022-09-15 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural analysis of PTPN21 reveals a dominant-negative effect of the FERM domain on its phosphatase activity.
Sci Adv, 10, 2024
|
|
8GXE
 
 | | PTPN21 FERM PTP complex | | Descriptor: | CHLORIDE ION, Tyrosine-protein phosphatase non-receptor type 21 | | Authors: | Chen, L, Zheng, Y.Y, Zhou, C. | | Deposit date: | 2022-09-19 | | Release date: | 2023-09-27 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural analysis of PTPN21 reveals a dominant-negative effect of the FERM domain on its phosphatase activity.
Sci Adv, 10, 2024
|
|
4ZRI
 
 | | Crystal structure of Merlin-FERM and Lats2 | | Descriptor: | Merlin, Serine/threonine-protein kinase LATS2 | | Authors: | Li, F, Zhou, H, Long, J, Shen, Y. | | Deposit date: | 2015-05-12 | | Release date: | 2015-06-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Angiomotin binding-induced activation of Merlin/NF2 in the Hippo pathway
Cell Res., 25, 2015
|
|
4ZRJ
 
 | | Structure of Merlin-FERM and CTD | | Descriptor: | GLYCEROL, Merlin | | Authors: | Lin, Z, Li, F, Long, J, Shen, Y. | | Deposit date: | 2015-05-12 | | Release date: | 2015-06-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Angiomotin binding-induced activation of Merlin/NF2 in the Hippo pathway
Cell Res., 25, 2015
|
|
4RMA
 
 | | Crystal structure of the FERM domain of human ezrin | | Descriptor: | Ezrin, SULFATE ION | | Authors: | Phang, J.M, Harrop, S.J, Duff, A.P, Wilk, K.E, Curmi, P.M.G. | | Deposit date: | 2014-10-21 | | Release date: | 2015-12-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural characterization suggests models for monomeric and dimeric forms of full-length ezrin.
Biochem. J., 473, 2016
|
|
4RM8
 
 | | Crystal structure of human ezrin in space group P21 | | Descriptor: | Ezrin | | Authors: | Phang, J.M, Harrop, S.J, Davies, R, Duff, A.P, Wilk, K.E, Curmi, P.M.G. | | Deposit date: | 2014-10-20 | | Release date: | 2015-12-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural characterization suggests models for monomeric and dimeric forms of full-length ezrin.
Biochem. J., 473, 2016
|
|
4RM9
 
 | | Crystal structure of human ezrin in space group C2221 | | Descriptor: | Ezrin | | Authors: | Phang, J.M, Harrop, S.J, Davies, R, Duff, A.P, Wilk, K.E, Curmi, P.M.G. | | Deposit date: | 2014-10-21 | | Release date: | 2015-12-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural characterization suggests models for monomeric and dimeric forms of full-length ezrin.
Biochem. J., 473, 2016
|
|
4ZRK
 
 | | Merlin-FERM and Lats1 complex | | Descriptor: | Merlin, Serine/threonine-protein kinase LATS1 | | Authors: | Lin, Z, Li, Y, Wei, Z, Zhang, M. | | Deposit date: | 2015-05-12 | | Release date: | 2015-06-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.316 Å) | | Cite: | Angiomotin binding-induced activation of Merlin/NF2 in the Hippo pathway
Cell Res., 25, 2015
|
|
2I1J
 
 | | Moesin from Spodoptera frugiperda at 2.1 angstroms resolution | | Descriptor: | CHLORIDE ION, GLYCEROL, Moesin, ... | | Authors: | Li, Q, Nance, M.R, Tesmer, J.J.G. | | Deposit date: | 2006-08-14 | | Release date: | 2006-12-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Self-masking in an Intact ERM-merlin Protein: An Active Role for the Central alpha-Helical Domain.
J.Mol.Biol., 365, 2007
|
|
2I1K
 
 | |
