1D1N
 
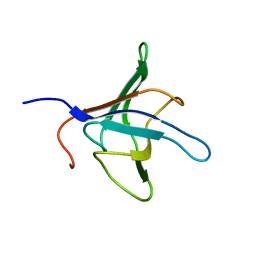 | | SOLUTION STRUCTURE OF THE FMET-TRNAFMET BINDING DOMAIN OF BECILLUS STEAROTHERMOPHILLUS TRANSLATION INITIATION FACTOR IF2 | | Descriptor: | INITIATION FACTOR 2 | | Authors: | Meunier, S, Spurio, S, Czisch, M, Wechselberger, R, Gueunneugues, M. | | Deposit date: | 1999-09-20 | | Release date: | 2000-09-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the fMet-tRNA(fMet)-binding domain of B. stearothermophilus initiation factor IF2.
EMBO J., 19, 2000
|
|
3E1Y
 
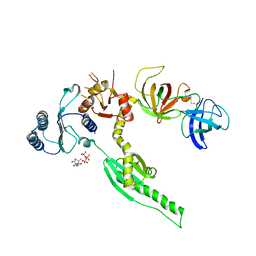 | | Crystal structure of human eRF1/eRF3 complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Eukaryotic peptide chain release factor GTP-binding subunit ERF3A, Eukaryotic peptide chain release factor subunit 1 | | Authors: | Cheng, Z, Lim, M, Kong, C, Song, H. | | Deposit date: | 2008-08-05 | | Release date: | 2009-05-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural insights into eRF3 and stop codon recognition by eRF1
Genes Dev., 23, 2009
|
|
1PN6
 
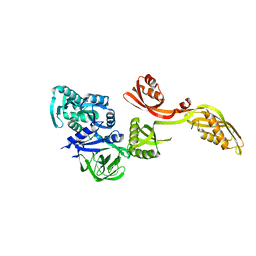 | | Domain-wise fitting of the crystal structure of T.thermophilus EF-G into the low resolution map of the release complex.Puromycin.EFG.GDPNP of E.coli 70S ribosome. | | Descriptor: | Elongation factor G | | Authors: | Valle, M, Zavialov, A, Sengupta, J, Rawat, U, Ehrenberg, M, Frank, J. | | Deposit date: | 2003-06-12 | | Release date: | 2003-07-15 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (10.8 Å) | | Cite: | Locking and Unlocking of Ribosomal Motions
Cell(Cambridge,Mass.), 114, 2003
|
|
7QTT
 
 | |
4MYU
 
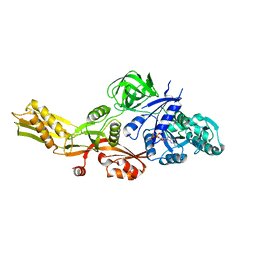 | |
8B6Z
 
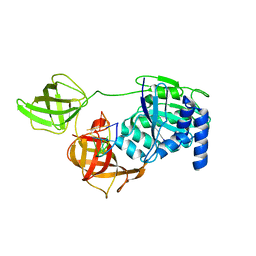 | |
3MMP
 
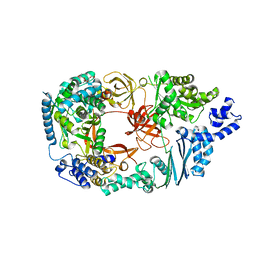 | | Structure of the Qb replicase, an RNA-dependent RNA polymerase consisting of viral and host proteins | | Descriptor: | (2S)-1-[3-{[(2R)-2-hydroxypropyl]oxy}-2,2-bis({[(2R)-2-hydroxypropyl]oxy}methyl)propoxy]propan-2-ol, Elongation factor Tu 2, Elongation factor Ts, ... | | Authors: | Kidmose, R.T, Vasiliev, N.N, Chetverin, A.B, Knudsen, C.R, Andersen, G.R. | | Deposit date: | 2010-04-20 | | Release date: | 2010-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the Qbeta replicase, an RNA-dependent RNA polymerase consisting of viral and host proteins.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2RDO
 
 | | 50S subunit with EF-G(GDPNP) and RRF bound | | Descriptor: | 23S RIBOSOMAL RNA, 50S ribosomal protein L1, 50S ribosomal protein L11, ... | | Authors: | Gao, N, Zavialov, A.V, Ehrenberg, M, Frank, J. | | Deposit date: | 2007-09-24 | | Release date: | 2008-03-04 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (9.1 Å) | | Cite: | Specific interaction between EF-G and RRF and its implication for GTP-dependent ribosome splitting into subunits.
J.Mol.Biol., 374, 2007
|
|
5FG3
 
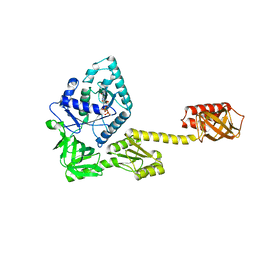 | | Crystal structure of GDP-bound aIF5B from Aeropyrum pernix | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Probable translation initiation factor IF-2 | | Authors: | Murakami, R, Miyoshi, T, Uchiumi, T, Ito, K. | | Deposit date: | 2015-12-20 | | Release date: | 2016-05-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of translation initiation factor 5B from the crenarchaeon Aeropyrum pernix.
Proteins, 84, 2016
|
|
5TY0
 
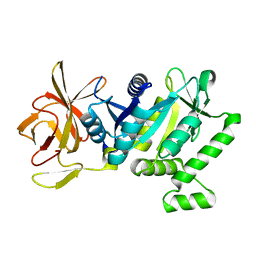 | | 2.22 Angstrom Crystal Structure of N-terminal Fragment (residues 1-419) of Elongation Factor G from Legionella pneumophila. | | Descriptor: | Elongation factor G, SODIUM ION, beta-D-glucopyranose | | Authors: | Minasov, G, Wawrzak, Z, Shuvalova, L, Cardona-Correa, A, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-11-17 | | Release date: | 2016-11-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | 2.22 Angstrom Crystal Structure of N-terminal Fragment (residues 1-419) of Elongation Factor G from Legionella pneumophila.
To Be Published
|
|
6BK7
 
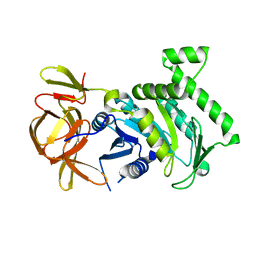 | | 1.83 Angstrom Resolution Crystal Structure of N-terminal Fragment (residues 1-404) of Elongation Factor G from Enterococcus faecalis | | Descriptor: | Elongation factor G, SODIUM ION | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Cardona-Correa, A, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-11-07 | | Release date: | 2017-11-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | 1.83 Angstrom Resolution Crystal Structure of N-terminal Fragment (residues 1-404) of Elongation Factor G from Enterococcus faecalis.
To be Published
|
|
6D9J
 
 | | Mammalian 80S ribosome with a double translocated CrPV-IRES, P-sitetRNA and eRF1. | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S12, ... | | Authors: | Pisareva, V.P, Pisarev, A.V, Fernandez, I.S. | | Deposit date: | 2018-04-30 | | Release date: | 2018-06-06 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Dual tRNA mimicry in the Cricket Paralysis Virus IRES uncovers an unexpected similarity with the Hepatitis C Virus IRES.
Elife, 7, 2018
|
|
7LS2
 
 | | 80S ribosome from mouse bound to eEF2 (Class I) | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | Authors: | Loerch, S, Smith, P.R, Kunder, N, Stanowick, A.D, Lou, T.-F, Campbell, Z.T. | | Deposit date: | 2021-02-17 | | Release date: | 2021-11-03 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Functionally distinct roles for eEF2K in the control of ribosome availability and p-body abundance.
Nat Commun, 12, 2021
|
|
7LS1
 
 | | 80S ribosome from mouse bound to eEF2 (Class II) | | Descriptor: | 28S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Loerch, S, Smith, P.R, Kunder, N, Stanowick, A.D, Lou, T.-F, Campbell, Z.T. | | Deposit date: | 2021-02-17 | | Release date: | 2021-11-03 | | Last modified: | 2021-12-08 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Functionally distinct roles for eEF2K in the control of ribosome availability and p-body abundance.
Nat Commun, 12, 2021
|
|
9L5T
 
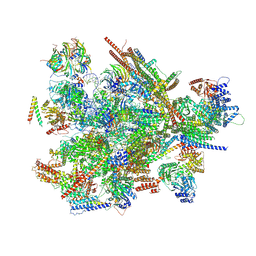 | | Cryo-EM structure of the thermophile spliceosome (state B*Q2) | | Descriptor: | Anaphase-promoting complex subunit 4-like WD40 domain-containing protein, CCDC12, Delta(14)-sterol reductase, ... | | Authors: | Li, Y, Fischer, P, Wang, M, Yuan, R, Meng, W, Luehrmann, R, Lau, B, Hurt, E, Cheng, J. | | Deposit date: | 2024-12-23 | | Release date: | 2025-03-12 | | Last modified: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural insights into spliceosome fidelity: DHX35-GPATCH1- mediated rejection of aberrant splicing substrates.
Cell Res., 35, 2025
|
|
9L5S
 
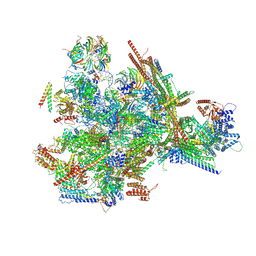 | | Cryo-EM structure of the thermophile spliceosome (state B*Q1) | | Descriptor: | Anaphase-promoting complex subunit 4-like WD40 domain-containing protein, CCDC12, Delta(14)-sterol reductase, ... | | Authors: | Li, Y, Fischer, P, Wang, M, Yuan, R, Meng, W, Luehrmann, R, Lau, B, Hurt, E, Cheng, J. | | Deposit date: | 2024-12-23 | | Release date: | 2025-03-12 | | Last modified: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into spliceosome fidelity: DHX35-GPATCH1- mediated rejection of aberrant splicing substrates.
Cell Res., 35, 2025
|
|
9L5R
 
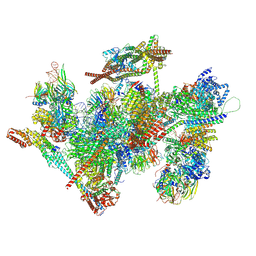 | | Cryo-EM structure of the thermophile spliceosome (state ILS) | | Descriptor: | Anaphase-promoting complex subunit 4-like WD40 domain-containing protein, CCDC12, Cell cycle control protein, ... | | Authors: | Li, Y, Fischer, P, Wang, M, Yuan, R, Meng, W, Luehrmann, R, Lau, B, Hurt, E, Cheng, J. | | Deposit date: | 2024-12-23 | | Release date: | 2025-03-12 | | Last modified: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insights into spliceosome fidelity: DHX35-GPATCH1- mediated rejection of aberrant splicing substrates.
Cell Res., 35, 2025
|
|
8G6J
 
 | | mRNA decoding in human is kinetically and structurally distinct from bacteria (GA state 2) | | Descriptor: | (3R,6R,9S,12S,15S,18S,20R,24aR)-6-[(2S)-butan-2-yl]-3,12-bis[(1R)-1-hydroxy-2-methylpropyl]-8,9,11,17,18-pentamethyl-15-[(2S)-2-methylbutyl]hexadecahydropyrido[1,2-a][1,4,7,10,13,16,19]heptaazacyclohenicosine-1,4,7,10,13,16,19(21H)-heptone, (3beta)-O~3~-[(2R)-2,6-dihydroxy-2-(2-methoxy-2-oxoethyl)-6-methylheptanoyl]cephalotaxine, 1,4-DIAMINOBUTANE, ... | | Authors: | Holm, M, Natchiar, K.S, Rundlet, E.J, Myasnikov, A.G, Altman, R.B, Blanchard, S.C. | | Deposit date: | 2023-02-15 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | mRNA decoding in human is kinetically and structurally distinct from bacteria.
Nature, 617, 2023
|
|
8G5Z
 
 | | mRNA decoding in human is kinetically and structurally distinct from bacteria (GA state) | | Descriptor: | 1,4-DIAMINOBUTANE, 18S rRNA, 28S rRNA, ... | | Authors: | Holm, M, Natchiar, K.S, Rundlet, E.J, Myasnikov, A.G, Altman, R.B, Blanchard, S.C. | | Deposit date: | 2023-02-14 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | mRNA decoding in human is kinetically and structurally distinct from bacteria.
Nature, 617, 2023
|
|
8G60
 
 | | mRNA decoding in human is kinetically and structurally distinct from bacteria (CR state) | | Descriptor: | 1,4-DIAMINOBUTANE, 18S rRNA, 28S rRNA, ... | | Authors: | Holm, M, Natchiar, K.S, Rundlet, E.J, Myasnikov, A.G, Altman, R.B, Blanchard, S.C. | | Deposit date: | 2023-02-14 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | mRNA decoding in human is kinetically and structurally distinct from bacteria.
Nature, 617, 2023
|
|
7F67
 
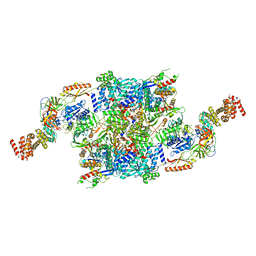 | | eIF2B-SFSV NSs-2-eIF2 | | Descriptor: | Eukaryotic translation initiation factor 2 subunit 1, Eukaryotic translation initiation factor 2 subunit 3, Non-structural protein NS-S, ... | | Authors: | Kashiwagi, K, Ito, T. | | Deposit date: | 2021-06-24 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | eIF2B-capturing viral protein NSs suppresses the integrated stress response.
Nat Commun, 12, 2021
|
|
7F66
 
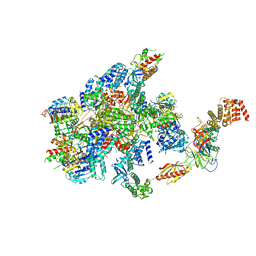 | | eIF2B-SFSV NSs-1-eIF2 | | Descriptor: | Eukaryotic translation initiation factor 2 subunit 1, Eukaryotic translation initiation factor 2 subunit 3, Non-structural protein NS-S, ... | | Authors: | Kashiwagi, K, Ito, T. | | Deposit date: | 2021-06-24 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | eIF2B-capturing viral protein NSs suppresses the integrated stress response.
Nat Commun, 12, 2021
|
|
7DVQ
 
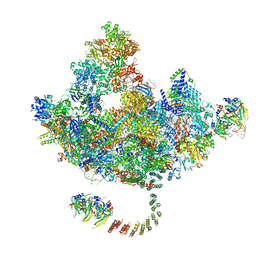 | | Cryo-EM Structure of the Activated Human Minor Spliceosome (minor Bact Complex) | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, 5'-O-[(S)-hydroxy{[(R)-hydroxy{[(S)-hydroxy(methoxy)phosphoryl]oxy}phosphoryl]oxy}phosphoryl]guanosine, Armadillo repeat-containing protein 7, ... | | Authors: | Bai, R, Wan, R, Wang, L, Xu, K, Zhang, Q, Lei, J, Shi, Y. | | Deposit date: | 2021-01-14 | | Release date: | 2021-03-31 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structure of the activated human minor spliceosome.
Science, 371, 2021
|
|
1XB2
 
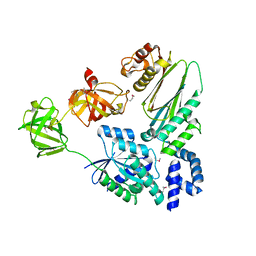 | | Crystal Structure of Bos taurus mitochondrial Elongation Factor Tu/Ts Complex | | Descriptor: | Elongation factor Ts, mitochondrial, Elongation factor Tu | | Authors: | Jeppesen, M.G, Navratil, T, Spremulli, L.L, Nyborg, J. | | Deposit date: | 2004-08-27 | | Release date: | 2004-11-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Bovine Mitochondrial Elongation Factor Tu.Ts Complex
J.Biol.Chem., 280, 2005
|
|
1ZM3
 
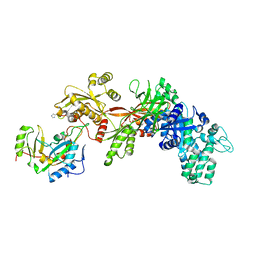 | | Structure of the apo eEF2-ETA complex | | Descriptor: | Elongation factor 2, exotoxin A | | Authors: | Joergensen, R, Merrill, A.R, Yates, S.P, Marquez, V.E, Schwan, A.L, Boesen, T, Andersen, G.R. | | Deposit date: | 2005-05-10 | | Release date: | 2005-05-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | Exotoxin A-eEF2 complex structure indicates ADP ribosylation by ribosome mimicry.
Nature, 436, 2005
|
|
