4MOD
 
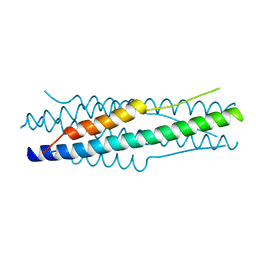 | | Structure of the MERS-CoV fusion core | | Descriptor: | HR1 of S protein, LINKER, HR2 of S protein | | Authors: | Gao, J, Lu, G, Qi, J, Li, Y, Wu, Y, Deng, Y, Geng, H, Xiao, H, Tan, W, Yan, J, Gao, G.F. | | Deposit date: | 2013-09-12 | | Release date: | 2013-10-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Structure of the fusion core and inhibition of fusion by a heptad repeat peptide derived from the S protein of Middle East respiratory syndrome coronavirus.
J.Virol., 87, 2013
|
|
1ZVA
 
 | | A structure-based mechanism of SARS virus membrane fusion | | Descriptor: | E2 glycoprotein | | Authors: | Deng, Y, Liu, J, Zheng, Q, Yong, W, Dai, J, Lu, M. | | Deposit date: | 2005-06-01 | | Release date: | 2006-05-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structures and Polymorphic Interactions of Two Heptad-Repeat Regions of the SARS Virus S2 Protein.
Structure, 14, 2006
|
|
1WNC
 
 | | Crystal structure of the SARS-CoV Spike protein fusion core | | Descriptor: | E2 glycoprotein | | Authors: | Xu, Y, Lou, Z, Liu, Y, Pang, H, Tien, P, Gao, G.F, Rao, Z. | | Deposit date: | 2004-07-29 | | Release date: | 2004-09-07 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of severe acute respiratory syndrome coronavirus spike protein fusion core
J.Biol.Chem., 279, 2004
|
|
2IEQ
 
 | |
6M3W
 
 | | Post-fusion structure of SARS-CoV spike glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Fan, X, Cao, D, Zhang, X. | | Deposit date: | 2020-03-04 | | Release date: | 2020-06-03 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM analysis of the post-fusion structure of the SARS-CoV spike glycoprotein.
Nat Commun, 11, 2020
|
|
5ZHY
 
 | | Structural characterization of the HCoV-229E fusion core | | Descriptor: | Spike glycoprotein | | Authors: | Zhang, W, Zheng, Q, Yan, M, Chen, X, Yang, H, Zhou, W, Rao, Z. | | Deposit date: | 2018-03-13 | | Release date: | 2018-06-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.441 Å) | | Cite: | Structural characterization of the HCoV-229E fusion core.
Biochem. Biophys. Res. Commun., 497, 2018
|
|
6B3O
 
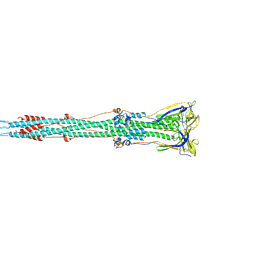 | | Tectonic conformational changes of a coronavirus spike glycoprotein promote membrane fusion | | Descriptor: | Spike glycoprotein | | Authors: | Walls, A.C, Tortorici, M.A, Snijder, J, Xiong, X, Bosch, B.J, Rey, F.A, Veesler, D. | | Deposit date: | 2017-09-22 | | Release date: | 2017-10-04 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Tectonic conformational changes of a coronavirus spike glycoprotein promote membrane fusion.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8U1G
 
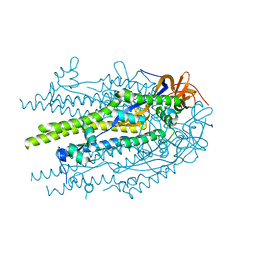 | | Prefusion-stabilized SARS-CoV-2 S2 subunit | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S2 | | Authors: | Hsieh, C.-L, McLellan, J.S. | | Deposit date: | 2023-08-31 | | Release date: | 2024-01-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Prefusion-stabilized SARS-CoV-2 S2-only antigen provides protection against SARS-CoV-2 challenge.
Nat Commun, 15, 2024
|
|
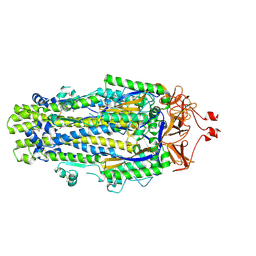
8VQB
 
 | |
8VQA
 
 | |
8DYA
 
 | |
8FDW
 
 | | Cryo-EM structure of SARS-CoV-2 postfusion spike in membrane | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S2, ... | | Authors: | Zhang, J, Shi, W, Cai, Y.F, Zhu, H.S, Peng, H.Q, Voyer, J, Volloch, S.R, Cao, H, Mayer, M.L, Song, K.K, Xu, C, Lu, J.M, Chen, B. | | Deposit date: | 2022-12-05 | | Release date: | 2023-05-10 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structure of SARS-CoV-2 postfusion spike in membrane.
Nature, 619, 2023
|
|
7WXZ
 
 | | Crystal structure of the recombinant protein HR121 from the S2 protein of SARS-CoV-2 | | Descriptor: | Spike protein S2' | | Authors: | Zheng, Y.T, Ouyang, S, Pang, W, Lu, Y, Zhao, Y.B. | | Deposit date: | 2022-02-15 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | A variant-proof SARS-CoV-2 vaccine targeting HR1 domain in S2 subunit of spike protein.
Cell Res., 32, 2022
|
|
7E9T
 
 | | Nanometer resolution in situ structure of SARS-CoV-2 post-fusion spike | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S2, ... | | Authors: | Zhu, Y, Tai, L, Zhu, G, Yin, G, Sun, F. | | Deposit date: | 2021-03-05 | | Release date: | 2021-11-17 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (10.9 Å) | | Cite: | Nanometer-resolution in situ structure of the SARS-CoV-2 postfusion spike protein.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7COT
 
 | |
8VAO
 
 | |
8VCR
 
 | | SARS-CoV-2 Spike S2 bound to Fab 54043-5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 54043-5 Fab Heavy Chain, ... | | Authors: | Johnson, N.V, McLellan, J.S. | | Deposit date: | 2023-12-14 | | Release date: | 2024-07-31 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Discovery and characterization of a pan-betacoronavirus S2-binding antibody.
Structure, 32, 2024
|
|
7Y9N
 
 | | an engineered 5-helix bundle derived from SARS-CoV-2 S2 in complex with HR2P | | Descriptor: | SARS-coV-2 S2 subunit, Spike protein S2',5HB-H2 | | Authors: | Lu, G.W, Lin, X, Guo, L.Y, Lin, S. | | Deposit date: | 2022-06-25 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.885 Å) | | Cite: | An engineered 5-helix bundle derived from SARS-CoV-2 S2 pre-binds sarbecoviral spike at both serological- and endosomal-pH to inhibit virus entry.
Emerg Microbes Infect, 11, 2022
|
|
8VQ9
 
 | |
7ZR2
 
 | | Crystal structure of a chimeric protein mimic of SARS-CoV-2 Spike HR1 in complex with HR2 | | Descriptor: | Spike protein S2', Spike protein S2',Chimeric protein mimic of SARS-CoV-2 Spike HR1 | | Authors: | Camara-Artigas, A, Gavira, J.A, Cano-Munoz, M, Polo-Megias, D, Conejero-Lara, F. | | Deposit date: | 2022-05-03 | | Release date: | 2022-11-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Novel chimeric proteins mimicking SARS-CoV-2 spike epitopes with broad inhibitory activity.
Int.J.Biol.Macromol., 222, 2022
|
|
8V5V
 
 | | Structure of a SARS-CoV-2 spike S2 subunit in a pre-fusion, open conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S2,Fibritin, ... | | Authors: | Olmedillas, E, Diaz, R, Hastie, K, Ollmann-Saphire, E. | | Deposit date: | 2023-12-01 | | Release date: | 2024-12-18 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structure of a SARS-CoV-2 spike S2 subunit in a pre-fusion, open conformation
To Be Published
|
|
7LRT
 
 | |
5X5F
 
 | | Prefusion structure of MERS-CoV spike glycoprotein, conformation 2 | | Descriptor: | S protein | | Authors: | Yuan, Y, Cao, D, Zhang, Y, Ma, J, Qi, J, Wang, Q, Lu, G, Wu, Y, Yan, J, Shi, Y, Zhang, X, Gao, G.F. | | Deposit date: | 2017-02-15 | | Release date: | 2017-05-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
5I08
 
 | | Prefusion structure of a human coronavirus spike protein | | Descriptor: | Spike glycoprotein,Foldon chimera | | Authors: | Kirchdoerfer, R.N, Cottrell, C.A, Wang, N, Pallesen, J, Yassine, H.M, Turner, H.L, Corbett, K.S, Graham, B.S, McLellan, J.S, Ward, A.B. | | Deposit date: | 2016-02-03 | | Release date: | 2016-03-02 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.04 Å) | | Cite: | Pre-fusion structure of a human coronavirus spike protein.
Nature, 531, 2016
|
|
3JCL
 
 | | Cryo-electron microscopy structure of a coronavirus spike glycoprotein trimer | | Descriptor: | Spike glycoprotein | | Authors: | Walls, A.C, Tortorici, M.A, Bosch, B.J, Frenz, B, Rottier, P.J.M, DiMaio, F, Rey, F.A, Veesler, D. | | Deposit date: | 2015-12-21 | | Release date: | 2016-02-03 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-electron microscopy structure of a coronavirus spike glycoprotein trimer.
Nature, 531, 2016
|
|
