7MYX
 
 | |
7VS3
 
 | | The crystal structure of rat calcium-dependent activator protein for secretion (CAPS) C2PH | | Descriptor: | Calcium-dependent secretion activator 1, SULFATE ION | | Authors: | Zhou, H, Wei, Z.Q, Zhang, L, Ren, Y.J, Ma, C. | | Deposit date: | 2021-10-25 | | Release date: | 2023-02-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.595 Å) | | Cite: | The C 2 and PH domains of CAPS constitute an effective PI(4,5)P2-binding unit essential for Ca 2+ -regulated exocytosis.
Structure, 31, 2023
|
|
6F24
 
 | | PH domain from PfAPH | | Descriptor: | C-terminal PH domain from PfAPH | | Authors: | Darvill, N, Liu, B, Matthews, S, Soldati-Favre, D, Rouse, S, Benjamin, S, Blake, T, Dubois, D.J, Hammoudi, P.M, Pino, P. | | Deposit date: | 2017-11-23 | | Release date: | 2018-12-12 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | C-terminal PH domain from P. falciparum acylated plekstrin homology domain containing protein (APH)
To Be Published
|
|
2P0H
 
 | | ArhGAP9 PH domain in complex with Ins(1,3,4)P3 | | Descriptor: | (1S,3S,4S)-1,3,4-TRIPHOSPHO-MYO-INOSITOL, Rho GTPase-activating protein 9 | | Authors: | Ceccarelli, D.F.J, Blasutig, I, Goudreault, M, Ruston, J, Pawson, T, Sicheri, F. | | Deposit date: | 2007-02-28 | | Release date: | 2007-03-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Non-canonical Interaction of Phosphoinositides with Pleckstrin Homology Domains of Tiam1 and ArhGAP9.
J.Biol.Chem., 282, 2007
|
|
2P0D
 
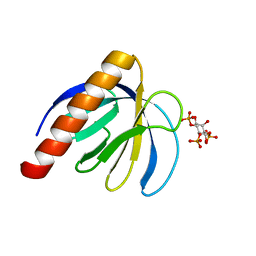 | | ArhGAP9 PH domain in complex with Ins(1,4,5)P3 | | Descriptor: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Rho GTPase-activating protein 9 | | Authors: | Ceccarelli, D.F.J, Blasutig, I, Goudreault, M, Ruston, J, Pawson, T, Sicheri, F. | | Deposit date: | 2007-02-28 | | Release date: | 2007-03-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.811 Å) | | Cite: | Non-canonical Interaction of Phosphoinositides with Pleckstrin Homology Domains of Tiam1 and ArhGAP9.
J.Biol.Chem., 282, 2007
|
|
5MR1
 
 | | Crystal structure of the Pleckstrin homology domain of Interactor protein for cytohesin exchange factors 1 (IPCEF1) | | Descriptor: | Interactor protein for cytohesin exchange factors 1 | | Authors: | Newman, J.A, Aitkenhead, H, Wang, D, Burgess-Brown, N, Williams, E, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2016-12-21 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of the Pleckstrin homology domain of Interactor protein for cytohesin exchange factors 1 (IPCEF1)
To be published
|
|
8OZZ
 
 | |
7YIR
 
 | |
7YIS
 
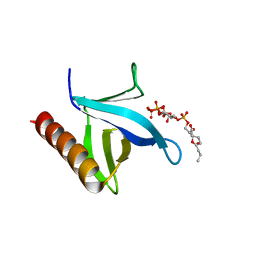 | | Crystal structure of N-terminal PH domain of ARAP3 protein in complex with inositol 1,3,4,5-tetrakisphosphate | | Descriptor: | (2R)-3-{[(S)-{[(2S,3R,5S,6S)-2,6-DIHYDROXY-3,4,5-TRIS(PHOSPHONOOXY)CYCLOHEXYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-2-(1-HYDROXY BUTOXY)PROPYL BUTYRATE, Arf-GAP with Rho-GAP domain, ANK repeat and PH domain-containing protein 3 | | Authors: | Zhang, Y.J, Liu, Y.R, Wu, B. | | Deposit date: | 2022-07-18 | | Release date: | 2023-05-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural Insights Uncover the Specific Phosphoinositide Recognition by the PH1 Domain of Arap3.
Int J Mol Sci, 24, 2023
|
|
7KJZ
 
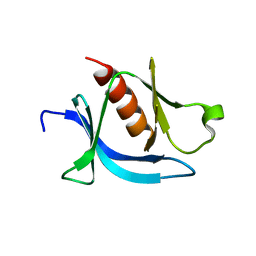 | | crystal structure of PLEKHA7 PH domain biding inositol-tetraphosphate | | Descriptor: | 1,2-ETHANEDIOL, INOSITOL-(1,3,4,5)-TETRAKISPHOSPHATE, Pleckstrin homology domain-containing family A member 7 | | Authors: | Marassi, F.M, Aleshin, A.E, Liddington, R.C. | | Deposit date: | 2020-10-26 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural basis for the association of PLEKHA7 with membrane-embedded phosphatidylinositol lipids.
Structure, 29, 2021
|
|
7KK7
 
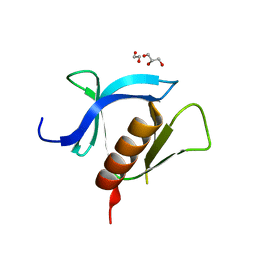 | | crystal structure of ligand-free PLEKHA7 PH domain | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Pleckstrin homology domain-containing family A member 7 | | Authors: | Marassi, F.M, Aleshin, A.E, Liddington, R.C. | | Deposit date: | 2020-10-27 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for the association of PLEKHA7 with membrane-embedded phosphatidylinositol lipids.
Structure, 29, 2021
|
|
7KJO
 
 | |
5C79
 
 | | PH domain of ASAP1 in complex with diC4-PtdIns(4,5)P2 | | Descriptor: | (2R)-3-{[(R)-HYDROXY{[(1R,2R,3S,4R,5R,6S)-2,3,6-TRIHYDROXY-4,5-BIS(PHOSPHONOOXY)CYCLOHEXYL]OXY}PHOSPHORYL]OXY}PROPANE-1 ,2-DIYL DIBUTANOATE, Arf-GAP, CHLORIDE ION | | Authors: | Xia, D, Tang, W.K. | | Deposit date: | 2015-06-24 | | Release date: | 2015-10-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular Basis for Cooperative Binding of Anionic Phospholipids to the PH Domain of the Arf GAP ASAP1.
Structure, 23, 2015
|
|
5U78
 
 | | Crystal structure of ORP8 PH domain in P1211 space group | | Descriptor: | Oxysterol-binding protein-related protein 8 | | Authors: | Ghai, R, Yang, H. | | Deposit date: | 2016-12-12 | | Release date: | 2017-10-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.978 Å) | | Cite: | ORP5 and ORP8 bind phosphatidylinositol-4, 5-biphosphate (PtdIns(4,5)P 2) and regulate its level at the plasma membrane.
Nat Commun, 8, 2017
|
|
5C6R
 
 | | Crystal structure of PH domain of ASAP1 | | Descriptor: | Arf-GAP, PHOSPHATE ION, TRIETHYLENE GLYCOL | | Authors: | Xia, D, Tang, W.K. | | Deposit date: | 2015-06-23 | | Release date: | 2015-10-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular Basis for Cooperative Binding of Anionic Phospholipids to the PH Domain of the Arf GAP ASAP1.
Structure, 23, 2015
|
|
5U77
 
 | | Crystal structure of ORP8 PH domain | | Descriptor: | FORMIC ACID, N-(2-hydroxyethyl)-N,N-dimethyl-3-sulfopropan-1-aminium, Oxysterol-binding protein-related protein 8 | | Authors: | Ghai, R, Yang, H. | | Deposit date: | 2016-12-11 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.157 Å) | | Cite: | ORP5 and ORP8 bind phosphatidylinositol-4, 5-biphosphate (PtdIns(4,5)P 2) and regulate its level at the plasma membrane.
Nat Commun, 8, 2017
|
|
1P6S
 
 | | Solution Structure of the Pleckstrin Homology Domain of Human Protein Kinase B beta (Pkb/Akt) | | Descriptor: | RAC-beta serine/threonine protein kinase | | Authors: | Auguin, D, Barthe, P, Auge-Senegas, M.T, Stern, M.H, Noguchi, M, Roumestand, C. | | Deposit date: | 2003-04-30 | | Release date: | 2004-05-18 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Solution structure and backbone dynamics of the pleckstrin homology domain of the human
protein kinase B (PKB/Akt). Interaction with inositol phosphates.
J.BIOMOL.NMR, 28, 2004
|
|
1PLS
 
 | | SOLUTION STRUCTURE OF A PLECKSTRIN HOMOLOGY DOMAIN | | Descriptor: | PLECKSTRIN HOMOLOGY DOMAIN | | Authors: | Yoon, H.S, Hajduk, P.J, Petros, A.M, Olejniczak, E.T, Meadows, R.P, Fesik, S.W. | | Deposit date: | 1994-05-03 | | Release date: | 1995-06-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a pleckstrin-homology domain.
Nature, 369, 1994
|
|
2DYN
 
 | | DYNAMIN (PLECKSTRIN HOMOLOGY DOMAIN) (DYNPH) | | Descriptor: | DYNAMIN | | Authors: | Timm, D.E. | | Deposit date: | 1997-07-21 | | Release date: | 1997-11-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the pleckstrin homology domain from dynamin.
Nat.Struct.Biol., 1, 1994
|
|
2I5F
 
 | | Crystal structure of the C-terminal PH domain of pleckstrin in complex with D-myo-Ins(1,2,3,5,6)P5 | | Descriptor: | (1R,2R,3R,4R,5S,6S)-6-HYDROXYCYCLOHEXANE-1,2,3,4,5-PENTAYL PENTAKIS[DIHYDROGEN (PHOSPHATE)], Pleckstrin | | Authors: | Jackson, S.G, Haslam, R.J, Junop, M.S. | | Deposit date: | 2006-08-24 | | Release date: | 2007-08-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural analysis of the carboxy terminal PH domain of pleckstrin bound to D-myo-inositol 1,2,3,5,6-pentakisphosphate.
Bmc Struct.Biol., 7, 2007
|
|
2I5C
 
 | | Crystal structure of the C-terminal PH domain of pleckstrin in complex with D-myo-Ins(1,2,3,4,5)P5 | | Descriptor: | (1R,2S,3R,4S,5S,6R)-6-HYDROXYCYCLOHEXANE-1,2,3,4,5-PENTAYL PENTAKIS[DIHYDROGEN (PHOSPHATE)], Pleckstrin | | Authors: | Jackson, S.G, Haslam, R.J, Junop, M.S. | | Deposit date: | 2006-08-24 | | Release date: | 2007-08-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural analysis of the carboxy terminal PH domain of pleckstrin bound to D-myo-inositol 1,2,3,5,6-pentakisphosphate.
Bmc Struct.Biol., 7, 2007
|
|
3PP2
 
 | | Crystal structure of the pleckstrin homology domain of ArhGAP27 | | Descriptor: | CITRIC ACID, GLYCEROL, Rho GTPase-activating protein 27, ... | | Authors: | Shen, L, Tempel, W, Tong, Y, Nedyalkova, L, Li, Y, Wernimont, A.K, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-11-23 | | Release date: | 2010-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.421 Å) | | Cite: | Crystal structure of the pleckstrin homology domain of ArhGAP27
to be published
|
|
1BAK
 
 | |
3NSU
 
 | | A Systematic Screen for Protein-Lipid Interactions in Saccharomyces cerevisiae | | Descriptor: | Phosphatidylinositol 4,5-bisphosphate-binding protein SLM1, SULFATE ION | | Authors: | Gallego, O, Fernandez-Tornero, C, Aguilar-Gurrieri, C, Muller, C, Gavin, A.C. | | Deposit date: | 2010-07-02 | | Release date: | 2010-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A systematic screen for protein-lipid interactions in Saccharomyces cerevisiae.
Mol. Syst. Biol., 6, 2010
|
|
1DRO
 
 | | NMR STRUCTURE OF THE CYTOSKELETON/SIGNAL TRANSDUCTION PROTEIN | | Descriptor: | BETA-SPECTRIN | | Authors: | Zhang, P, Talluri, S, Deng, H, Branton, D, Wagner, G. | | Deposit date: | 1995-09-29 | | Release date: | 1996-04-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the pleckstrin homology domain of Drosophila beta-spectrin.
Structure, 3, 1995
|
|
