4Z0I
 
 | | Crystal structure of a tetramer of GluA2 ligand binding domains bound with glutamate at 1.45 Angstrom resolution | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLUTAMIC ACID, Glutamate receptor 2,Glutamate receptor 2, ... | | Authors: | Baranovic, J, Chebli, M, Salazar, H, Carbone, A.L, Ghisi, V, Faelber, K, Lau, A.Y, Daumke, O, Plested, A.J.R. | | Deposit date: | 2015-03-26 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of the tetrameric wt GluA2 ligand-binding domain bound to glutamate at 1.45 Angstroms resolution
To Be Published
|
|
2CMO
 
 | | The structure of a mixed glur2 ligand-binding core dimer in complex with (s)-glutamate and the antagonist (s)-ns1209 | | Descriptor: | 2-({[(3E)-5-{4-[(DIMETHYLAMINO)(DIHYDROXY)-LAMBDA~4~-SULFANYL]PHENYL}-8-METHYL-2-OXO-6,7,8,9-TETRAHYDRO-1H-PYRROLO[3,2-H]ISOQUINOLIN-3(2H)-YLIDENE]AMINO}OXY)-4-HYDROXYBUTANOIC ACID, GLUTAMATE RECEPTOR 2, GLUTAMIC ACID, ... | | Authors: | Kasper, C, Pickering, D.S, Mirza, O, Olsen, L, Kristensen, A.S, Greenwood, J.R, Liljefors, T, Schousboe, A, Watjen, F, Gajhede, M, Sigurskjold, B.W, Kastrup, J.S. | | Deposit date: | 2006-05-11 | | Release date: | 2006-06-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The Structure of a Mixed Glur2 Ligand-Binding Core Dimer in Complex with (S)-Glutamate and the Antagonist (S)-Ns1209.
J.Mol.Biol., 357, 2006
|
|
2A5S
 
 | | Crystal Structure Of The NR2A Ligand Binding Core In Complex With Glutamate | | Descriptor: | GLUTAMIC ACID, N-methyl-D-aspartate receptor NMDAR2A subunit | | Authors: | Furukawa, H, Singh, S.K, Mancusso, R, Gouaux, E. | | Deposit date: | 2005-06-30 | | Release date: | 2005-11-15 | | Last modified: | 2017-07-26 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Subunit arrangement and function in NMDA receptors
Nature, 438, 2005
|
|
6O9G
 
 | | Open state GluA2 in complex with STZ and blocked by AgTx-636, after micelle signal subtraction | | Descriptor: | CYCLOTHIAZIDE, GLUTAMIC ACID, Glutamate receptor 2,Voltage-dependent calcium channel gamma-2 subunit, ... | | Authors: | Twomey, E.C, Yelshanskaya, M.V, Vassilevski, A.A, Sobolevsky, A.I. | | Deposit date: | 2019-03-13 | | Release date: | 2019-03-20 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Mechanisms of Channel Block in Calcium-Permeable AMPA Receptors.
Neuron, 99, 2018
|
|
3T9V
 
 | |
3T9X
 
 | |
3T96
 
 | | Iodowillardiine bound to a double cysteine mutant (A452C/S652C) of the ligand binding domain of GluA2 | | Descriptor: | 2-AMINO-3-(5-IODO-2,4-DIOXO-3,4-DIHYDRO-2H-PYRIMIDIN-1-YL)-PROPIONIC ACID, Glutamate receptor 2, ZINC ION | | Authors: | Ahmed, A.H, Wang, S, Chuang, H.H, Oswald, R.E. | | Deposit date: | 2011-08-02 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.872 Å) | | Cite: | Mechanism of AMPA Receptor Activation by Partial Agonists: DISULFIDE TRAPPING OF CLOSED LOBE CONFORMATIONS.
J.Biol.Chem., 286, 2011
|
|
6ODL
 
 | | Crystal structure of GluN2A agonist binding domain with 4-butyl-(S)-CCG-IV | | Descriptor: | (1S,2R)-2-[(S)-amino(carboxy)methyl]-1-butylcyclopropane-1-carboxylic acid, Glutamate receptor ionotropic, NMDA 2A,Glutamate receptor ionotropic, ... | | Authors: | Mou, T.C, Clausen, R.P, Sprang, S.R, Hansen, K.B. | | Deposit date: | 2019-03-26 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Stereoselective synthesis of novel 2'-(S)-CCG-IV analogues as potent NMDA receptor agonists.
Eur.J.Med.Chem., 212, 2021
|
|
1WVJ
 
 | | Exploring the GluR2 ligand-binding core in complex with the bicyclic AMPA analogue (S)-4-AHCP | | Descriptor: | 3-(3-HYDROXY-7,8-DIHYDRO-6H-CYCLOHEPTA[D]ISOXAZOL-4-YL)-L-ALANINE, GLYCEROL, SULFATE ION, ... | | Authors: | Nielsen, B.B, Pickering, D.S, Greenwood, J.R, Brehm, L, Gajhede, M, Schousboe, A, Kastrup, J.S. | | Deposit date: | 2004-12-15 | | Release date: | 2005-04-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Exploring the GluR2 ligand-binding core in complex with the bicyclical AMPA analogue (S)-4-AHCP
FEBS J., 272, 2005
|
|
3T9H
 
 | | Kainate bound to a double cysteine mutant (A452C/S652C) of the ligand binding domain of GluA2 | | Descriptor: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, Glutamate receptor 2, ZINC ION | | Authors: | Ahmed, A.H, Wang, S, Chuang, H.H, Oswald, R.E. | | Deposit date: | 2011-08-02 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.015 Å) | | Cite: | Mechanism of AMPA Receptor Activation by Partial Agonists: DISULFIDE TRAPPING OF CLOSED LOBE CONFORMATIONS.
J.Biol.Chem., 286, 2011
|
|
2F34
 
 | |
6OVD
 
 | | Crystal structure of GluN1/GluN2A NMDA receptor agonist binding domains with glycine and antagonist, 3-ethylphenyl-ACEPC | | Descriptor: | (3S,5S)-5-[(2R)-2-amino-2-carboxyethyl]-1-(3-ethylphenyl)pyrazolidine-3-carboxylic acid, GLYCINE, Glutamate receptor ionotropic, ... | | Authors: | Syrenne, J.T, Mou, T.C, Tamborini, L, Pinto, A, Sprang, S.R, Hansen, K.B. | | Deposit date: | 2019-05-07 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Crystal structure of GluN1/GluN2A NMDA receptor agonist binding domains with glycine and antagonist, 3-ethylphenyl-ACEPC
To Be Published
|
|
3T93
 
 | |
2F35
 
 | |
3TDJ
 
 | | Crystal structure of the GluA2 ligand-binding domain (S1S2J-L483Y-N754S) in complex with glutamate and BPAM-97 at 1.95 A resolution | | Descriptor: | 4-ethyl-7-fluoro-3,4-dihydro-2H-1,2,4-benzothiadiazine 1,1-dioxide, CHLORIDE ION, GLUTAMIC ACID, ... | | Authors: | Krintel, C, Frydenvang, K, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2011-08-11 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Thermodynamics and structural analysis of positive allosteric modulation of the ionotropic glutamate receptor GluA2.
Biochem.J., 441, 2012
|
|
3T9U
 
 | |
3UA8
 
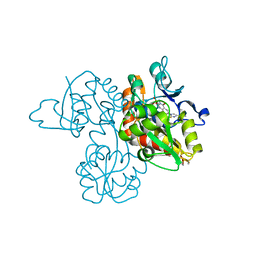 | | Crystal Structure Analysis of a 6-Amino Quinazolinedione Sulfonamide bound to human GluR2 | | Descriptor: | Glutamate receptor 2, N-methyl-1-{3-[(methylsulfonyl)amino]-2,4-dioxo-7-(trifluoromethyl)-1,2,3,4-tetrahydroquinazolin-6-yl}-1H-imidazole-4-carboxamide | | Authors: | Kallen, J. | | Deposit date: | 2011-10-21 | | Release date: | 2012-01-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 6-Amino quinazolinedione sulfonamides as orally active competitive AMPA receptor antagonists.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
6OVE
 
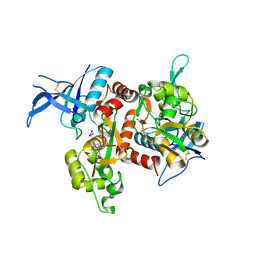 | | Crystal structure of GluN1/GluN2A NMDA receptor agonist binding domains with glycine and antagonist, 4-propylphenyl-ACEPC | | Descriptor: | (3R,5S)-5-[(2R)-2-amino-2-carboxyethyl]-1-(4-propylphenyl)pyrazolidine-3-carboxylic acid, GLYCINE, Glutamate receptor ionotropic, ... | | Authors: | Syrenne, J.T, Mou, T.C, Tamborini, L, Pinto, A, Sprang, S.R, Hansen, K.B. | | Deposit date: | 2019-05-07 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of GluN1/GluN2A NMDA receptor agonist binding domains with glycine and antagonist, 3-ethylphenyl-ACEPC
To Be Published
|
|
5CBR
 
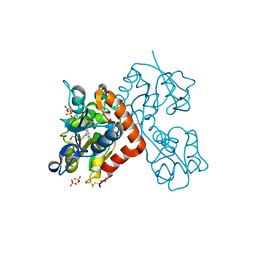 | | Crystal structure of the GluA2 ligand-binding domain (S1S2J) in complex with the antagonist (S)-2-amino-3-(3,4-dichloro-5-(5-hydroxypyridin-3-yl)phenyl)propanoic acid at 2.0A resolution | | Descriptor: | 3,4-dichloro-5-(5-hydroxypyridin-3-yl)-L-phenylalanine, ACETATE ION, GLYCEROL, ... | | Authors: | Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2015-07-01 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.996 Å) | | Cite: | Studies on Aryl-Substituted Phenylalanines: Synthesis, Activity, and Different Binding Modes at AMPA Receptors.
J.Med.Chem., 59, 2016
|
|
5CC2
 
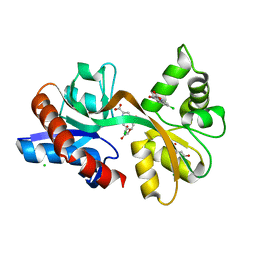 | | STRUCTURE OF THE LIGAND-BINDING DOMAIN OF THE IONOTROPIC GLUTAMATE RECEPTOR-LIKE GLUD2 IN COMPLEX WITH 7-CKA | | Descriptor: | 7-Chlorokynurenic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Naur, P, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2015-07-01 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Pharmacology and Structural Analysis of Ligand Binding to the Orthosteric Site of Glutamate-Like GluD2 Receptors.
Mol.Pharmacol., 89, 2016
|
|
6PEQ
 
 | |
5CBS
 
 | | Crystal structure of the GluA2 ligand-binding domain (S1S2J) in complex with the antagonist (R)-2-amino-3-(3'-hydroxybiphenyl-3-yl)propanoic acid at 1.8A resolution | | Descriptor: | (R)-2-amino-3-(3'-hydroxybiphenyl-3-yl)propanoic acid, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2015-07-01 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Studies on Aryl-Substituted Phenylalanines: Synthesis, Activity, and Different Binding Modes at AMPA Receptors.
J.Med.Chem., 59, 2016
|
|
5CMM
 
 | |
5CMK
 
 | | Crystal structure of the GluK2EM LBD dimer assembly complex with glutamate and LY466195 | | Descriptor: | (3S,4aR,6S,8aR)-6-{[(2S)-2-carboxy-4,4-difluoropyrrolidin-1-yl]methyl}decahydroisoquinoline-3-carboxylic acid, CHLORIDE ION, GLUTAMIC ACID, ... | | Authors: | Chittori, S, Mayer, M.L. | | Deposit date: | 2015-07-16 | | Release date: | 2016-07-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structural basis of kainate subtype glutamate receptor desensitization.
Nature, 537, 2016
|
|
5DEX
 
 | | Crystal structure of GluN1/GluN2A NMDA receptor agonist binding domains with glycine and antagonist, phenyl-ACEPC | | Descriptor: | 5-[(2R)-2-amino-2-carboxyethyl]-1-phenyl-1H-pyrazole-3-carboxylic acid, GLYCINE, Glutamate receptor ionotropic, ... | | Authors: | Mou, T.-C, Conti, P, Pinto, A, Tamborini, L, Sprang, S.R, Hansen, K.B. | | Deposit date: | 2015-08-26 | | Release date: | 2016-09-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of subunit selectivity for competitive NMDA receptor antagonists with preference for GluN2A over GluN2B subunits.
Proc. Natl. Acad. Sci. U.S.A., 2017
|
|
