3M4O
 
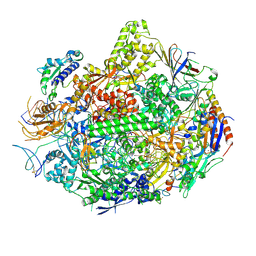 | | RNA polymerase II elongation complex B | | Descriptor: | DNA (28-MER), DNA (5'-D(P*GP*TP*GP*GP*TP*TP*AP*TP*GP*GP*GP*TP*AP*G)-3'), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Wang, D, Zhu, G, Huang, X, Lippard, S.J. | | Deposit date: | 2010-03-11 | | Release date: | 2010-05-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.57 Å) | | Cite: | X-ray structure and mechanism of RNA polymerase II stalled at an antineoplastic monofunctional platinum-DNA adduct.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
6H67
 
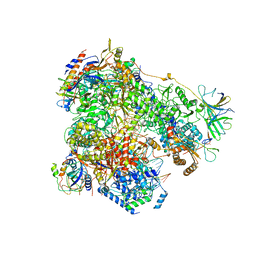 | | Yeast RNA polymerase I elongation complex stalled by cyclobutane pyrimidine dimer (CPD) | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Sanz-Murillo, M, Xu, J, Gil-Carton, D, Wang, D, Fernandez-Tornero, C. | | Deposit date: | 2018-07-26 | | Release date: | 2018-08-29 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of RNA polymerase I stalling at UV light-induced DNA damage.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
8TVW
 
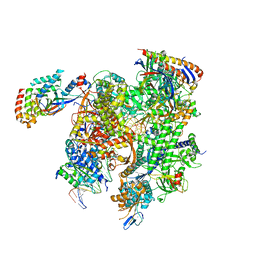 | | Cryo-EM structure of CPD-stalled Pol II (conformation 1) | | Descriptor: | DNA (NTS), DNA (TS), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Sarsam, R.D, Lahiri, I, Leschziner, A.E. | | Deposit date: | 2023-08-18 | | Release date: | 2024-01-24 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Elf1 promotes Rad26's interaction with lesion-arrested Pol II for transcription-coupled repair.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
4BBS
 
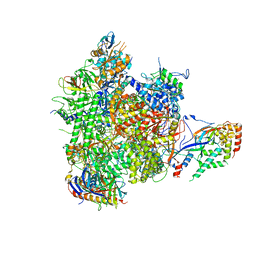 | | Structure of an initially transcribing RNA polymerase II-TFIIB complex | | Descriptor: | 5'-D(*AP*GP*CP*GP*CP*AP*GP*TP*TP*GP*TP*GP*CP*TP *AP*TP*GP*AP*TP*AP*TP*TP*TP*TP*TP*AP*TP)-3', 5'-D(*GP*GP*CP*AP*CP*AP*AP*CP*TP*GP*CP*GP*CP*TP)-3', 5'-R(*AP*UP*AP*UP*CP*AP)-3', ... | | Authors: | Sainsbury, S, Niesser, J, Cramer, P. | | Deposit date: | 2012-09-27 | | Release date: | 2012-11-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure and Function of the Initially Transcribing RNA Polymerase II-TFIIB Complex
Nature, 493, 2013
|
|
6UQ1
 
 | | RNA polymerase II elongation complex with 5-guanidinohydantoin lesion in state 6 | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Oh, J, Wang, D. | | Deposit date: | 2019-10-18 | | Release date: | 2020-06-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | RNA polymerase II stalls on oxidative DNA damage via a torsion-latch mechanism involving lone pair-pi and CH-pi interactions.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8UKU
 
 | | RNA polymerase II elongation complex with Fapy-dG lesion with CMP added | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Hou, P, Oh, J, Wang, D. | | Deposit date: | 2023-10-15 | | Release date: | 2024-04-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Molecular Mechanism of RNA Polymerase II Transcriptional Mutagenesis by the Epimerizable DNA Lesion, Fapy·dG.
J.Am.Chem.Soc., 146, 2024
|
|
8UKT
 
 | | RNA polymerase II elongation complex with Fapy-dG lesion with AMP added | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Hou, P, Oh, J, Wang, D. | | Deposit date: | 2023-10-15 | | Release date: | 2024-04-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Molecular Mechanism of RNA Polymerase II Transcriptional Mutagenesis by the Epimerizable DNA Lesion, Fapy·dG.
J.Am.Chem.Soc., 146, 2024
|
|
3HOW
 
 | | Complete RNA polymerase II elongation complex III with a T-U mismatch and a frayed RNA 3'-uridine | | Descriptor: | 5'-D(*AP*CP*TP*AP*CP*TP*TP*GP*AP*GP*CP*T)-3', 5'-D(*AP*GP*CP*TP*C*AP*AP*GP*TP*AP*GP*TP*TP*AP*TP*GP*CP*CP*(BRU)P*GP*GP*TP*CP*AP*TP*T)-3', 5'-R(*UP*GP*CP*AP*UP*UP*U*CP*AP*AP*CP*CP*AP*GP*GP*CP*UP*U)-3', ... | | Authors: | Sydow, J.F, Brueckner, F, Cheung, A.C.M, Damsma, G.E, Dengl, S, Lehmann, E, Vassylyev, D, Cramer, P. | | Deposit date: | 2009-06-03 | | Release date: | 2009-07-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural basis of transcription: mismatch-specific fidelity mechanisms and paused RNA polymerase II with frayed RNA.
Mol.Cell, 34, 2009
|
|
4BY1
 
 | | elongating RNA Polymerase II-Bye1 TLD complex soaked with AMPCPP | | Descriptor: | 5'-D(*AP*AP*AP*GP*TP*AP*CP*TP*TP*GP*AP*GP*CP*TP)-3', 5'-D(*AP*GP*CP*TP*CP*AP*AP*GP*TP*AP*CP*TP*TP*AP *TP*TP*CP*CP*BRUP*GP*GP*TP*CP*AP*AP*T)-3', 5'-R(*UP*UP*CP*GP*AP*CP*CP*AP*GP*GP*AP)-3', ... | | Authors: | Kinkelin, K, Wozniak, G.G, Rothbart, S.B, Lidschreiber, M, Strahl, B.D, Cramer, P. | | Deposit date: | 2013-07-17 | | Release date: | 2013-09-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structures of RNA Polymerase II Complexes with Bye1, a Chromatin-Binding Phf3/Dido1 Homologue
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2NVX
 
 | | RNA polymerase II elongation complex in 5 mM Mg+2 with 2'-dUTP | | Descriptor: | 28-MER DNA template strand, 5'-D(*CP*TP*GP*CP*TP*TP*AP*TP*CP*GP*GP*TP*AP*G)-3', 5'-R(*AP*UP*CP*GP*AP*GP*AP*GP*GP*A)-3', ... | | Authors: | Wang, D, Bushnell, D.A, Westover, K.D, Kaplan, C.D, Kornberg, R.D. | | Deposit date: | 2006-11-13 | | Release date: | 2006-12-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis
Cell(Cambridge,Mass.), 127, 2006
|
|
8CEO
 
 | |
7KED
 
 | | RNA polymerase II elongation complex with unnatural base dTPT3 | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Oh, J, Wang, W, Wang, D. | | Deposit date: | 2020-10-10 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Transcriptional processing of an unnatural base pair by eukaryotic RNA polymerase II.
Nat.Chem.Biol., 17, 2021
|
|
5W4U
 
 | |
1SFO
 
 | | RNA POLYMERASE II STRAND SEPARATED ELONGATION COMPLEX | | Descriptor: | DNA STRAND, DNA-directed RNA polymerase II 13.6 kDa polypeptide, DNA-directed RNA polymerase II 14.2 kDa polypeptide, ... | | Authors: | Westover, K.D, Bushnell, D.A, Kornberg, R.D. | | Deposit date: | 2004-02-20 | | Release date: | 2004-03-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.61 Å) | | Cite: | Structural Basis of Transcription: Separation of RNA from DNA by RNA Polymerase II
Science, 303, 2004
|
|
3HOZ
 
 | | Complete RNA polymerase II elongation complex IV with a T-U mismatch and a frayed RNA 3'-guanine | | Descriptor: | 5'-D(*AP*CP*TP*AP*CP*TP*TP*GP*AP*GP*CP*T)-3', 5'-D(*AP*GP*CP*TP*C*AP*AP*GP*TP*AP*GP*TP*TP*CP*TP*GP*CP*CP*(BRU)P*GP*GP*TP*CP*AP*TP*T)-3', 5'-R(*UP*GP*CP*AP*UP*UP*U*CP*AP*AP*CP*CP*AP*GP*GP*CP*UP*G)-3', ... | | Authors: | Sydow, J.F, Brueckner, F, Cheung, A.C.M, Damsma, G.E, Dengl, S, Lehmann, E, Vassylyev, D, Cramer, P. | | Deposit date: | 2009-06-03 | | Release date: | 2009-07-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Structural basis of transcription: mismatch-specific fidelity mechanisms and paused RNA polymerase II with frayed RNA.
Mol.Cell, 34, 2009
|
|
3HOX
 
 | | Complete RNA polymerase II elongation complex V | | Descriptor: | 5'-D(*AP*CP*TP*AP*CP*TP*TP*GP*AP*GP*CP*T)-3', 5'-D(*AP*GP*CP*TP*C*AP*AP*GP*TP*AP*GP*TP*TP*AP*AP*GP*CP*CP*(BRU)P*GP*GP*TP*CP*AP*TP*T)-3', 5'-R(*UP*GP*CP*AP*UP*UP*U*CP*AP*AP*CP*CP*AP*GP*GP*CP*UP*U)-3', ... | | Authors: | Sydow, J.F, Brueckner, F, Cheung, A.C.M, Damsma, G.E, Dengl, S, Lehmann, E, Vassylyev, D, Cramer, P. | | Deposit date: | 2009-06-03 | | Release date: | 2009-07-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Structural basis of transcription: mismatch-specific fidelity mechanisms and paused RNA polymerase II with frayed RNA.
Mol.Cell, 34, 2009
|
|
8TVP
 
 | | Cryo-EM structure of CPD-stalled Pol II in complex with Rad26 (open state) | | Descriptor: | DNA (NTS), DNA (TS), DNA repair and recombination protein RAD26, ... | | Authors: | Sarsam, R.D, Lahiri, I, Leschziner, A.E. | | Deposit date: | 2023-08-18 | | Release date: | 2024-01-24 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Elf1 promotes Rad26's interaction with lesion-arrested Pol II for transcription-coupled repair.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8TVV
 
 | | Cryo-EM structure of backtracked Pol II | | Descriptor: | DNA (NTS), DNA (TS), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Sarsam, R.D, Lahiri, I, Leschziner, A.E. | | Deposit date: | 2023-08-18 | | Release date: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Elf1 promotes Rad26's interaction with lesion-arrested Pol II for transcription-coupled repair.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8TVX
 
 | | Cryo-EM structure of CPD-stalled Pol II (Conformation 2) | | Descriptor: | DNA (NTS), DNA (TS), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Sarsam, R.D, Lahiri, I, Leschziner, A.E. | | Deposit date: | 2023-08-18 | | Release date: | 2024-01-24 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Elf1 promotes Rad26's interaction with lesion-arrested Pol II for transcription-coupled repair.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
5C3E
 
 | | Crystal structure of a transcribing RNA Polymerase II complex reveals a complete transcription bubble | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Barnes, C.O, Calero, M, Malik, I, Spahr, H, Zhang, Q, Pullara, F, Kaplan, C.D, Calero, G. | | Deposit date: | 2015-06-17 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Crystal Structure of a Transcribing RNA Polymerase II Complex Reveals a Complete Transcription Bubble.
Mol.Cell, 59, 2015
|
|
7RIY
 
 | | RNA polymerase II elongation complex with hairpin polyamide Py-Im 1, scaffold 2 soaked with UTP | | Descriptor: | 3-({3-[(3-{[4-({4-[(4-{[4-({(2R)-2-amino-4-[(1-methyl-4-{[1-methyl-4-({1-methyl-4-[(1-methyl-1H-imidazole-2-carbonyl)amino]-1H-imidazole-2-carbonyl}amino)-1H-pyrrole-2-carbonyl]amino}-1H-pyrrole-2-carbonyl)amino]butanoyl}amino)-1-methyl-1H-imidazole-2-carbonyl]amino}-1-methyl-1H-pyrrole-2-carbonyl)amino]-1-methyl-1H-pyrrole-2-carbonyl}amino)-1-methyl-1H-pyrrole-2-carbonyl]amino}propyl)(methyl)amino]propyl}carbamoyl)benzoic acid, DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, ... | | Authors: | Oh, J, Dervan, P.B, Wang, D. | | Deposit date: | 2021-07-20 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | RNA polymerase II trapped on a molecular treadmill: Structural basis of persistent transcriptional arrest by a minor groove DNA binder.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6RQH
 
 | | RNA Polymerase I Closed Conformation 1 (CC1) | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Mueller, C.W, Sadian, Y, Tafur, L. | | Deposit date: | 2019-05-15 | | Release date: | 2019-12-11 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Molecular insight into RNA polymerase I promoter recognition and promoter melting.
Nat Commun, 10, 2019
|
|
6F40
 
 | | RNA Polymerase III open complex | | Descriptor: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Vorlaender, M.K, Khatter, H, Wetzel, R, Hagen, W.J.H, Mueller, C.W. | | Deposit date: | 2017-11-29 | | Release date: | 2018-01-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Molecular mechanism of promoter opening by RNA polymerase III.
Nature, 553, 2018
|
|
4A3J
 
 | | RNA Polymerase II initial transcribing complex with a 2nt DNA-RNA hybrid and soaked with GMPCPP | | Descriptor: | 5'-D(*AP*GP*CP*TP*AP*GP*CP*TP*TP*TP*CP*BRUP*AP*CP*CP *TP*GP*AP*AP*CP*AP*AP*CP*TP*AP*AP*CP)-3', 5'-D(*GP*TP*AP*GP*AP*AP*AP*GP*CP*TP*AP*GP*CP*TP)-3', 5'-R(*CP*AP)-3', ... | | Authors: | Cheung, A.C.M, Sainsbury, S, Cramer, P. | | Deposit date: | 2011-09-30 | | Release date: | 2011-12-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural Basis of Initial RNA Polymerase II Transcription.
Embo J., 30, 2011
|
|
8UOT
 
 | | Composite map of PICdeltaTFIIK form1 | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Yang, C, Murakami, K. | | Deposit date: | 2023-10-20 | | Release date: | 2024-10-23 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Transcription start site scanning requires the fungi-specific hydrophobic loop of Tfb3.
Nucleic Acids Res., 52, 2024
|
|
