3HOU
 
 | | Complete RNA polymerase II elongation complex I with a T-U mismatch | | Descriptor: | 5'-D(*A*AP*CP*TP*AP*CP*TP*TP*GP*AP*GP*CP*T)-3', 5'-D(*AP*GP*CP*TP*CP*A*AP*GP*TP*AP*GP*TP*TP*AP*TP*GP*CP*CP*(BRU)P*GP*GP*TP*CP*AP*TP*T)-3', 5'-R(*UP*GP*CP*AP*UP*U*UP*CP*GP*AP*CP*CP*AP*GP*GP*CP*U)-3', ... | | Authors: | Sydow, J.F, Brueckner, F, Cheung, A.C.M, Damsma, G.E, Dengl, S, Lehmann, E, Vassylyev, D, Cramer, P. | | Deposit date: | 2009-06-03 | | Release date: | 2009-07-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis of transcription: mismatch-specific fidelity mechanisms and paused RNA polymerase II with frayed RNA.
Mol.Cell, 34, 2009
|
|
7O4I
 
 | | Yeast RNA polymerase II transcription pre-initiation complex with initial transcription bubble | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Schilbach, S, Aibara, S, Dienemann, C, Grabbe, F, Cramer, P. | | Deposit date: | 2021-04-06 | | Release date: | 2021-06-16 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of RNA polymerase II pre-initiation complex at 2.9 angstrom defines initial DNA opening.
Cell, 184, 2021
|
|
7O75
 
 | | Yeast RNA polymerase II transcription pre-initiation complex with open promoter DNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Schilbach, S, Aibara, S, Dienemann, C, Grabbe, F, Cramer, P. | | Deposit date: | 2021-04-13 | | Release date: | 2021-06-16 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of RNA polymerase II pre-initiation complex at 2.9 angstrom defines initial DNA opening.
Cell, 184, 2021
|
|
8BWS
 
 | | Structure of yeast RNA Polymerase III elongation complex at 3.3 A | | Descriptor: | (3R,5S,7R,8R,9S,10S,12S,13R,14S,17R)-10,13-dimethyl-17-[(2R)-pentan-2-yl]-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthrene-3,7,12-triol, DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, ... | | Authors: | Nguyen, P.Q, Fernandez-Tornero, C. | | Deposit date: | 2022-12-07 | | Release date: | 2023-04-05 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of Ty1 integrase tethering to RNA polymerase III for targeted retrotransposon integration.
Nat Commun, 14, 2023
|
|
5OT2
 
 | | RNA polymerase II elongation complex in the presence of 3d-Napht-A | | Descriptor: | 2-ethyl-7-methoxy-naphthalene, DNA non-template strand, DNA template strand, ... | | Authors: | Malvezzi, S, Farnung, L, Aloisi, C, Angelov, T, Cramer, P, Sturla, S.J. | | Deposit date: | 2017-08-19 | | Release date: | 2017-11-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Mechanism of RNA polymerase II stalling by DNA alkylation.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6BLP
 
 | | Pol II elongation complex with an abasic lesion at i+1 position, soaking AMPCPP | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, DNA (5'-D(P*AP*(3DR)P*CP*TP*CP*TP*CP*GP*AP*TP*G)-3'), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Wang, W, Wang, D. | | Deposit date: | 2017-11-11 | | Release date: | 2018-02-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.203 Å) | | Cite: | Structural basis of transcriptional stalling and bypass of abasic DNA lesion by RNA polymerase II.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6HLS
 
 | | Yeast apo RNA polymerase I* | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Tafur, L, Sadian, Y, Weis, F, Muller, C.W. | | Deposit date: | 2018-09-11 | | Release date: | 2019-04-03 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | The cryo-EM structure of a 12-subunit variant of RNA polymerase I reveals dissociation of the A49-A34.5 heterodimer and rearrangement of subunit A12.2.
Elife, 8, 2019
|
|
3S16
 
 | | RNA Polymerase II Initiation Complex with an 8-nt RNA | | Descriptor: | DNA (5'-D(*CP*TP*AP*CP*CP*GP*AP*TP*AP*AP*GP*CP*AP*GP*AP*CP*GP*AP*TP*CP*CP*TP*CP*TP*CP*GP*AP*TP*G)-3'), DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, ... | | Authors: | Liu, X, Bushnell, D.A, Silva, D.A, Huang, X, Kornberg, R.D. | | Deposit date: | 2011-05-14 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.241 Å) | | Cite: | Initiation complex structure and promoter proofreading.
Science, 333, 2011
|
|
6TUT
 
 | | Cryo-EM structure of the RNA Polymerase III-Maf1 complex | | Descriptor: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Vorlaender, M.K, Hagen, W.J.H, Mueller, C.W. | | Deposit date: | 2020-01-08 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structural basis for RNA polymerase III transcription repression by Maf1.
Nat.Struct.Mol.Biol., 27, 2020
|
|
4C3H
 
 | | Structure of 14-subunit RNA polymerase I at 3.27 A resolution, crystal form C2-93 | | Descriptor: | DNA-DIRECTED RNA POLYMERASE I SUBUNIT RPA12, DNA-DIRECTED RNA POLYMERASE I SUBUNIT RPA135, DNA-DIRECTED RNA POLYMERASE I SUBUNIT RPA14, ... | | Authors: | Fernandez-Tornero, C, Moreno-Morcillo, M, Rashid, U.J, Taylor, N.M.I, Ruiz, F.M, Gruene, T, Legrand, P, Steuerwald, U, Muller, C.W. | | Deposit date: | 2013-08-24 | | Release date: | 2013-10-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.27 Å) | | Cite: | Crystal Structure of the 14-Subunit RNA Polymerase I
Nature, 502, 2013
|
|
4BXX
 
 | | Arrested RNA polymerase II-Bye1 complex | | Descriptor: | 5'-D(*AP*GP*CP*TP*AP*GP*CP*TP*TP*AP*CP*CP*TP*GP *GP*TP*GP* BRUP*TP*GP*CP*TP*CP*TP*AP*AP*DC)-3', 5'-D(*CP*CP*CP*CP*CP*CP*CP*CP*CP*CP*CP)-3', 5'-D(*GP*AP*GP*GP*TP*AP*AP*GP*CP*TP*AP*GP*CP*TP)-3', ... | | Authors: | Kinkelin, K, Wozniak, G.G, Rothbart, S.B, Lidschreiber, M, Strahl, B.D, Cramer, P. | | Deposit date: | 2013-07-16 | | Release date: | 2013-09-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.28 Å) | | Cite: | Structures of RNA Polymerase II Complexes with Bye1, a Chromatin-Binding Phf3/Dido1 Homologue
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
7RIP
 
 | | RNA polymerase II elongation complex with hairpin polyamide Py-Im 1, scaffold 1 soaked with CTP | | Descriptor: | 3-({3-[(3-{[4-({4-[(4-{[4-({(2R)-2-amino-4-[(1-methyl-4-{[1-methyl-4-({1-methyl-4-[(1-methyl-1H-imidazole-2-carbonyl)amino]-1H-imidazole-2-carbonyl}amino)-1H-pyrrole-2-carbonyl]amino}-1H-pyrrole-2-carbonyl)amino]butanoyl}amino)-1-methyl-1H-imidazole-2-carbonyl]amino}-1-methyl-1H-pyrrole-2-carbonyl)amino]-1-methyl-1H-pyrrole-2-carbonyl}amino)-1-methyl-1H-pyrrole-2-carbonyl]amino}propyl)(methyl)amino]propyl}carbamoyl)benzoic acid, DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, ... | | Authors: | Oh, J, Dervan, P.B, Wang, D. | | Deposit date: | 2021-07-20 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | RNA polymerase II trapped on a molecular treadmill: Structural basis of persistent transcriptional arrest by a minor groove DNA binder.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
4Y7N
 
 | | The Structure Insight into 5-Carboxycytosine Recognition by RNA Polymerase II during Transcription Elongation. | | Descriptor: | DNA (29-MER), DNA (5'-D(*CP*TP*GP*CP*TP*TP*AP*TP*CP*GP*GP*TP*AP*G)-3'), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Wang, L, Chong, J, Wang, D. | | Deposit date: | 2015-02-15 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Molecular basis for 5-carboxycytosine recognition by RNA polymerase II elongation complex.
Nature, 523, 2015
|
|
3RZD
 
 | | RNA Polymerase II Initiation Complex with a 5-nt RNA | | Descriptor: | DNA (5'-D(*CP*TP*AP*CP*CP*GP*AP*TP*AP*AP*GP*CP*AP*GP*AP*CP*GP*AP*TP*CP*CP*TP*CP*TP*CP*GP*AP*TP*G)-3'), DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, ... | | Authors: | Liu, X, Bushnell, D.A, Silva, D.A, Huang, X, Kornberg, R.D. | | Deposit date: | 2011-05-11 | | Release date: | 2011-08-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Initiation complex structure and promoter proofreading.
Science, 333, 2011
|
|
3S1Q
 
 | | RNA Polymerase II Initiation Complex with a 5-nt 3'-deoxy RNA soaked with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (5'-D(*CP*TP*AP*CP*CP*GP*AP*TP*AP*AP*GP*CP*AP*GP*AP*CP*GP*AP*TP*CP*CP*TP*CP*TP*CP*GP*AP*TP*G)-3'), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Liu, X, Bushnell, D.A, Silva, D.A, Huang, X, Kornberg, R.D. | | Deposit date: | 2011-05-16 | | Release date: | 2011-08-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Initiation complex structure and promoter proofreading.
Science, 333, 2011
|
|
3S15
 
 | | RNA Polymerase II Initiation Complex with a 7-nt RNA | | Descriptor: | DNA (5'-D(*CP*TP*AP*CP*CP*GP*AP*TP*AP*AP*GP*CP*AP*GP*AP*CP*GP*AP*TP*CP*CP*TP*CP*TP*CP*GP*AP*TP*G)-3'), DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, ... | | Authors: | Liu, X, Bushnell, D.A, Silva, D.A, Huang, X, Kornberg, R.D. | | Deposit date: | 2011-05-14 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Initiation complex structure and promoter proofreading.
Science, 333, 2011
|
|
3S2H
 
 | | RNA Polymerase II Initiation Complex with a 6-nt RNA containing a 2[prime]-iodo ATP | | Descriptor: | DNA (5'-D(*CP*TP*AP*CP*CP*GP*AP*TP*AP*AP*GP*CP*AP*GP*AP*CP*GP*AP*TP*CP*CP*TP*CP*TP*CP*GP*AP*TP*G)-3'), DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, ... | | Authors: | Liu, X, Bushnell, D.A, Silva, D.A, Huang, X, Kornberg, R.D. | | Deposit date: | 2011-05-16 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Initiation complex structure and promoter proofreading.
Science, 333, 2011
|
|
7UIO
 
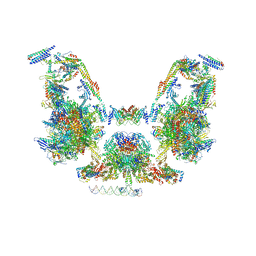 | | Mediator-PIC Early (Composite Model) | | Descriptor: | ALANINE, ASPARTIC ACID, DNA (37-MER), ... | | Authors: | Gorbea Colon, J.J, Chen, S.-F, Tsai, K.L, Murakami, K. | | Deposit date: | 2022-03-29 | | Release date: | 2023-03-01 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of a transcription pre-initiation complex on a divergent promoter.
Mol.Cell, 83, 2023
|
|
7UI9
 
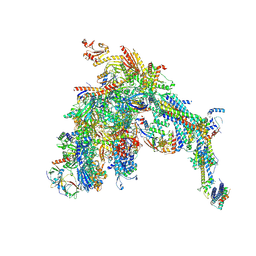 | | Core Mediator-PICearly (Copy A) | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Gorbea Colon, J.J, Chen, S.-F, Tsai, K.L, Murakami, K. | | Deposit date: | 2022-03-28 | | Release date: | 2023-02-15 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of a transcription pre-initiation complex on a divergent promoter.
Mol.Cell, 83, 2023
|
|
1TWG
 
 | | RNA polymerase II complexed with CTP | | Descriptor: | CYTIDINE-5'-TRIPHOSPHATE, DNA-directed RNA polymerase II 13.6 kDa polypeptide, DNA-directed RNA polymerase II 14.2 kDa polypeptide, ... | | Authors: | Westover, K.D, Bushnell, D.A, Kornberg, R.D. | | Deposit date: | 2004-06-30 | | Release date: | 2004-11-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis of transcription: nucleotide selection by rotation in the RNA polymerase II active center.
Cell(Cambridge,Mass.), 119, 2004
|
|
6EU3
 
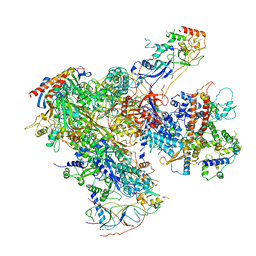 | | Apo RNA Polymerase III - closed conformation (cPOL3) | | Descriptor: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Abascal-Palacios, G, Ramsay, E.P, Beuron, F, Morris, E, Vannini, A. | | Deposit date: | 2017-10-27 | | Release date: | 2018-01-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of RNA polymerase III transcription initiation.
Nature, 553, 2018
|
|
3PO3
 
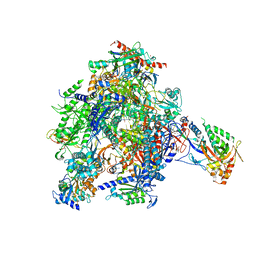 | | Arrested RNA Polymerase II reactivation intermediate | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, DNA non-template strand, ... | | Authors: | Cheung, A.C.M, Cramer, P. | | Deposit date: | 2010-11-21 | | Release date: | 2011-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis of RNA polymerase II backtracking, arrest and reactivation.
Nature, 471, 2011
|
|
3PO2
 
 | | Arrested RNA Polymerase II elongation complex | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Cheung, A.C.M, Cramer, P. | | Deposit date: | 2010-11-21 | | Release date: | 2011-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis of RNA polymerase II backtracking, arrest and reactivation.
Nature, 471, 2011
|
|
1I6H
 
 | | RNA POLYMERASE II ELONGATION COMPLEX | | Descriptor: | 5'-D(P*AP*AP*AP*TP*GP*CP*CP*TP*GP*GP*TP*CP*T)-3', 5'-R(P*GP*AP*CP*CP*AP*GP*GP*CP*A)-3', DNA-DIRECTED RNA POLYMERASE II 13.6KD POLYPEPTIDE, ... | | Authors: | Gnatt, A.L, Cramer, P, Kornberg, R.D. | | Deposit date: | 2001-03-02 | | Release date: | 2001-04-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis of transcription: an RNA polymerase II elongation complex at 3.3 A resolution.
Science, 292, 2001
|
|
6BQF
 
 | | Pol II elongation complex with 'dT-AP' at i+1, i-1 position | | Descriptor: | DNA (5'-D(P*CP*TP*(3DR)P*CP*TP*CP*TP*TP*GP*AP*TP*G)-3'), DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, ... | | Authors: | Wang, W, Wang, D. | | Deposit date: | 2017-11-27 | | Release date: | 2018-02-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Structural basis of transcriptional stalling and bypass of abasic DNA lesion by RNA polymerase II.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
