3FPU
 
 | |
2X6G
 
 | |
5COR
 
 | |
4RA8
 
 | | Structure analysis of the Mip1a P8A mutant | | Descriptor: | C-C motif chemokine 3 | | Authors: | Liang, W.G, Ren, M, Guo, Q, Tang, W.J. | | Deposit date: | 2014-09-09 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of human CCL18, CCL3, and CCL4 reveal molecular determinants for quaternary structures and sensitivity to insulin-degrading enzyme.
J.Mol.Biol., 427, 2015
|
|
7F1T
 
 | | Crystal structure of the human chemokine receptor CCR5 in complex with MIP-1a | | Descriptor: | C-C motif chemokine 3,C-C chemokine receptor type 5,Rubredoxin,C-C chemokine receptor type 5, ZINC ION | | Authors: | Zhang, H, Chen, K, Tan, Q, Han, S, Zhu, Y, Zhao, Q, Wu, B. | | Deposit date: | 2021-06-09 | | Release date: | 2021-07-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for chemokine recognition and receptor activation of chemokine receptor CCR5.
Nat Commun, 12, 2021
|
|
2X69
 
 | |
3KBX
 
 | |
4ZKB
 
 | | The chemokine binding protein of orf virus complexed with CCL3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C-C motif chemokine 3, Chemokine binding protein, ... | | Authors: | Knapp, K.M, Nakatani, Y, Krause, K.L. | | Deposit date: | 2015-04-30 | | Release date: | 2015-07-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of Orf Virus Chemokine Binding Protein in Complex with Host Chemokines Reveal Clues to Broad Binding Specificity.
Structure, 23, 2015
|
|
7F1Q
 
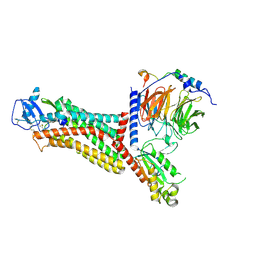 | | Cryo-EM structure of the chemokine receptor CCR5 in complex with MIP-1a and Gi | | Descriptor: | C-C motif chemokine 3,C-C chemokine receptor type 5, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhang, H, Chen, K, Tan, Q, Han, S, Zhu, Y, Zhao, Q, Wu, B. | | Deposit date: | 2021-06-09 | | Release date: | 2021-07-14 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for chemokine recognition and receptor activation of chemokine receptor CCR5.
Nat Commun, 12, 2021
|
|
3H44
 
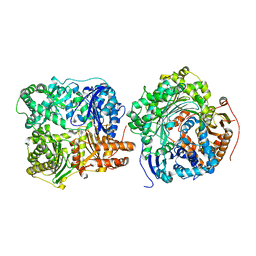 | |
5D65
 
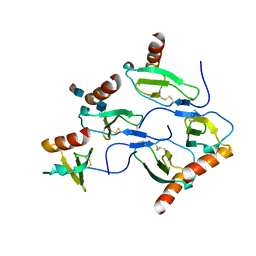 | | X-RAY STRUCTURE OF MACROPHAGE INFLAMMATORY PROTEIN-1 ALPHA (CCL3) WITH HEPARIN COMPLEX | | Descriptor: | 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid, C-C motif chemokine 3, CHLORIDE ION, ... | | Authors: | Liang, W.G, Hwang, D.Y, Zulueta, M.M, Hung, S.C, Tang, W. | | Deposit date: | 2015-08-11 | | Release date: | 2016-04-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for oligomerization and glycosaminoglycan binding of CCL5 and CCL3.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1B53
 
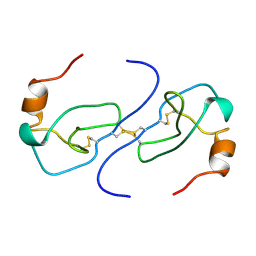 | | NMR STRUCTURE OF HUMAN MIP-1A D26A, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | MIP-1A | | Authors: | Waltho, J.P, Higgins, L.D, Craven, C.J, Tan, P, Dudgeon, T. | | Deposit date: | 1999-01-11 | | Release date: | 1999-07-22 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Identification of amino acid residues critical for aggregation of human CC chemokines macrophage inflammatory protein (MIP)-1alpha, MIP-1beta, and RANTES. Characterization of active disaggregated chemokine variants.
J.Biol.Chem., 274, 1999
|
|
1B50
 
 | | NMR STRUCTURE OF HUMAN MIP-1A D26A, 10 STRUCTURES | | Descriptor: | MIP-1A | | Authors: | Waltho, J.P, Higgins, L.D, Craven, C.J, Tan, P, Dudgeon, T. | | Deposit date: | 1999-01-11 | | Release date: | 1999-07-22 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Identification of amino acid residues critical for aggregation of human CC chemokines macrophage inflammatory protein (MIP)-1alpha, MIP-1beta, and RANTES. Characterization of active disaggregated chemokine variants.
J.Biol.Chem., 274, 1999
|
|
