7UYP
 
 | |
2HB5
 
 | | Crystal Structure of the Moloney Murine Leukemia Virus RNase H Domain | | Descriptor: | MAGNESIUM ION, Reverse transcriptase/ribonuclease H, SULFATE ION | | Authors: | Lim, D, Gregorio, G.G, Bingman, C.A, Martinez-Hackert, E, Hendrickson, W.A, Goff, S.P. | | Deposit date: | 2006-06-13 | | Release date: | 2006-08-29 | | Last modified: | 2023-03-22 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Crystal Structure of the Moloney Murine Leukemia Virus RNase H Domain.
J.Virol., 80, 2006
|
|
6MIH
 
 | | Crystal structure of host-guest complex with PC hachimoji DNA | | Descriptor: | DNA (5'-D(*CP*TP*TP*AP*(1WA)P*CP*(DB)P*T)-3'), DNA (5'-D(P*AP*(DS)P*GP*(1W5)P*TP*AP*AP*G)-3'), N-terminal fragment of MMLV reverse transcriptase | | Authors: | Georgiadis, M.M. | | Deposit date: | 2018-09-19 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Hachimoji DNA and RNA: A genetic system with eight building blocks.
Science, 363, 2019
|
|
7UYO
 
 | |
7UYN
 
 | |
8CS0
 
 | |
4XPC
 
 | |
6B1R
 
 | |
4XO0
 
 | |
6MIG
 
 | | Crystal structure of host-guest complex with PB hachimoji DNA | | Descriptor: | DNA (5'-D(*CP*TP*TP*AP*TP*(1WA)P*(1WA)P*(DS))-3'), DNA (5'-D(P*(DB)P*(1W5)P*(1W5)P*AP*TP*AP*AP*G)-3'), Gag-Pol polyprotein | | Authors: | Georgiadis, M.M. | | Deposit date: | 2018-09-19 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Hachimoji DNA and RNA: A genetic system with eight building blocks.
Science, 363, 2019
|
|
6MIK
 
 | | Crystal structure of host-guest complex with PP hachimoji DNA | | Descriptor: | DNA (5'-D(*CP*TP*TP*AP*TP*(1WA)P*(1WA)P*(DS))-3'), DNA (5'-D(P*(DB)P*(1W5)P*(1W5)P*AP*TP*AP*AP*G)-3'), N-terminal fragment of MMLV reverse transcriptase | | Authors: | Georgiadis, M.M. | | Deposit date: | 2018-09-19 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Hachimoji DNA and RNA: A genetic system with eight building blocks.
Science, 363, 2019
|
|
4M95
 
 | |
5VBS
 
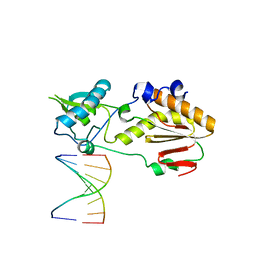 | | Structural basis for a six letter alphabet including GATCKX | | Descriptor: | DNA (5'-D(*CP*TP*TP*AP*TP*(DX)P*(DX)P*T)-3'), DNA (5'-D(P*AP*(93D)P*(93D)P*AP*TP*AP*AP*G)-3'), reverse transcriptase catalytic fragment | | Authors: | Singh, I, Georgiadis, M.M. | | Deposit date: | 2017-03-30 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.749 Å) | | Cite: | Structure and Biophysics for a Six Letter DNA Alphabet that Includes Imidazo[1,2-a]-1,3,5-triazine-2(8H)-4(3H)-dione (X) and 2,4-Diaminopyrimidine (K).
ACS Synth Biol, 6, 2017
|
|
3FSI
 
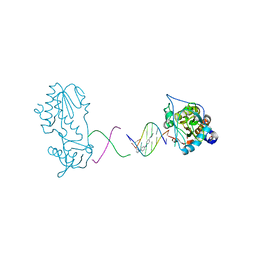 | | Crystal structure of a trypanocidal 4,4'-Bis(imidazolinylamino)diphenylamine bound to DNA | | Descriptor: | 5'-D(*CP*TP*TP*AP*AP*TP*TP*C)-3', 5'-D(P*GP*AP*AP*TP*TP*AP*AP*G)-3', ACETATE ION, ... | | Authors: | Glass, L.S, Georgiadis, M.M, Goodwin, K.D. | | Deposit date: | 2009-01-09 | | Release date: | 2009-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of a trypanocidal 4,4'-bis(imidazolinylamino)diphenylamine bound to DNA.
Biochemistry, 48, 2009
|
|
4XPE
 
 | |
2FJX
 
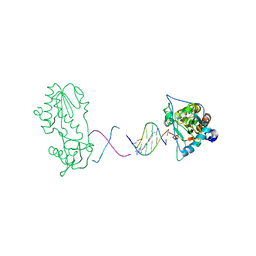 | | RT29 bound to D(CTTGAATGCATTCAAG) in complex with MMLV RT catalytic fragment | | Descriptor: | 2-(4-(4-CARBAMIMIDOYLPHENOXY)PHENYL)-1H-BENZO[D]IMIDAZOLE-6-CARBOXIMIDAMIDE, 5'-D(*CP*TP*TP*GP*AP*AP*TP*G)-3', 5'-D(P*CP*AP*TP*TP*CP*AP*AP*G)-3', ... | | Authors: | Goodwin, K.D, Georgiadis, M.M. | | Deposit date: | 2006-01-03 | | Release date: | 2006-06-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A High-Throughput, High-Resolution Strategy for the Study of Site-Selective DNA Binding Agents: Analysis of a "Highly Twisted" Benzimidazole-Diamidine.
J.Am.Chem.Soc., 128, 2006
|
|
1ZTW
 
 | |
1MML
 
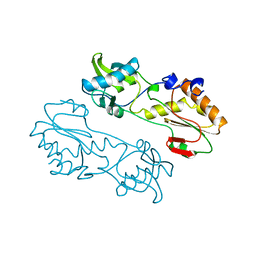 | | MECHANISTIC IMPLICATIONS FROM THE STRUCTURE OF A CATALYTIC FRAGMENT OF MMLV REVERSE TRANSCRIPTASE | | Descriptor: | MMLV REVERSE TRANSCRIPTASE | | Authors: | Georgiadis, M.M, Jessen, S.M, Ogata, C.M, Telesnitsky, A, Goff, S.P, Hendrickson, W.A. | | Deposit date: | 1995-07-18 | | Release date: | 1995-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanistic implications from the structure of a catalytic fragment of Moloney murine leukemia virus reverse transcriptase.
Structure, 3, 1995
|
|
8DW1
 
 | | Crystal structure of a host-guest complex with 5'-CTTAGTTATAACTAAG-3' | | Descriptor: | DNA (5'-D(*CP*TP*TP*AP*GP*TP*TP*A)-3'), DNA (5'-D(P*TP*AP*AP*CP*TP*AP*AP*G)-3'), reverse transcriptase | | Authors: | Georgiadis, M.M. | | Deposit date: | 2022-07-30 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.849 Å) | | Cite: | Two distinct rotations of bithiazole DNA intercalation revealed by direct comparison of crystal structures of Co(III)•bleomycin A 2 and B 2 bound to duplex 5'-TAGTT sites.
Bioorg.Med.Chem., 77, 2023
|
|
1ZTT
 
 | | Netropsin bound to d(CTTAATTCGAATTAAG) in complex with MMLV RT catalytic fragment | | Descriptor: | 5'-D(*CP*TP*TP*AP*AP*TP*TP*C)-3', 5'-D(P*GP*AP*AP*TP*TP*AP*AP*G)-3', NETROPSIN, ... | | Authors: | Goodwin, K.D, Long, E.C, Georgiadis, M.M. | | Deposit date: | 2005-05-27 | | Release date: | 2005-08-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A host-guest approach for determining drug-DNA interactions: an example using netropsin.
Nucleic Acids Res., 33, 2005
|
|
6GZA
 
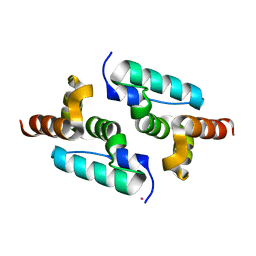 | | Structure of murine leukemia virus capsid C-terminal domain | | Descriptor: | COBALT (II) ION, Capsid protein | | Authors: | Qu, K, Dolezal, M, Schur, F.K.M, Murciano, B, Rumlova, M, Ruml, T, Briggs, J.A.G. | | Deposit date: | 2018-07-03 | | Release date: | 2018-12-05 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structure and architecture of immature and mature murine leukemia virus capsids.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6B1Q
 
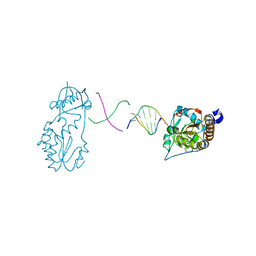 | |
2FJW
 
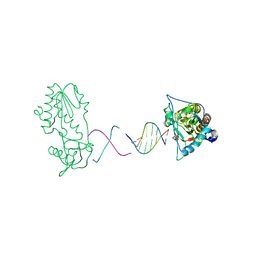 | | d(CTTGAATGCATTCAAG) in complex with MMLV RT catalytic fragment | | Descriptor: | 5'-D(*CP*TP*TP*GP*AP*AP*TP*G)-3', 5'-D(P*CP*AP*TP*TP*CP*AP*AP*G)-3', Reverse transcriptase | | Authors: | Goodwin, K.D, Georgiadis, M.M. | | Deposit date: | 2006-01-03 | | Release date: | 2006-06-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A High-Throughput, High-Resolution Strategy for the Study of Site-Selective DNA Binding Agents: Analysis of a "Highly Twisted" Benzimidazole-Diamidine.
J.Am.Chem.Soc., 128, 2006
|
|
6B1S
 
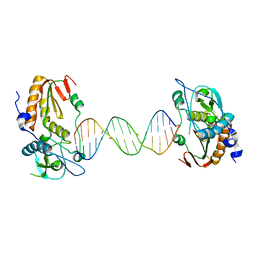 | |
1I6J
 
 | |
