1S32
 
 | | Molecular Recognition of the Nucleosomal 'Supergroove' | | Descriptor: | 2-(2-CARBAMOYLMETHOXY-ETHOXY)-ACETAMIDE, 3-AMINO-(DIMETHYLPROPYLAMINE), 4-AMINO-(1-METHYLIMIDAZOLE)-2-CARBOXYLIC ACID, ... | | Authors: | Edayathumangalam, R.S, Weyermann, P, Gottesfeld, J.M, Dervan, P.B, Luger, K. | | Deposit date: | 2004-01-12 | | Release date: | 2004-05-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Molecular Recognition of the Nucleosomal 'Supergroove'
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1M19
 
 | | LIGAND BINDING ALTERS THE STRUCTURE AND DYNAMICS OF NUCLEOSOMAL DNA | | Descriptor: | 3-AMINO-(DIMETHYLPROPYLAMINE), 4-AMINO-(1-METHYLIMIDAZOLE)-2-CARBOXYLIC ACID, 4-AMINO-(1-METHYLPYRROLE)-2-CARBOXYLIC ACID, ... | | Authors: | Suto, R.K, Edayathumangalam, R.S, White, C.L, Melander, C, Gottesfeld, J.M, Dervan, P.B, Luger, K. | | Deposit date: | 2002-06-18 | | Release date: | 2003-02-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of Nucleosome Core Particles in Complex with Minor Groove DNA-binding Ligands
J.Mol.Biol., 326, 2003
|
|
8T9F
 
 | |
1M1A
 
 | | LIGAND BINDING ALTERS THE STRUCTURE AND DYNAMICS OF NUCLEOSOMAL DNA | | Descriptor: | 3-AMINO-(DIMETHYLPROPYLAMINE), 4-AMINO-(1-METHYLIMIDAZOLE)-2-CARBOXYLIC ACID, 4-AMINO-(1-METHYLPYRROLE)-2-CARBOXYLIC ACID, ... | | Authors: | Suto, R.K, Edayathumangalam, R.S, White, C.L, Melander, C, Gottesfeld, J.M, Dervan, P.B, Luger, K. | | Deposit date: | 2002-06-18 | | Release date: | 2003-02-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal Structures of Nucleosome Core Particles in Complex with Minor Groove DNA-binding Ligands
J.MOL.BIOL., 326, 2003
|
|
8TOF
 
 | | Rpd3S bound to an H3K36Cme3 modified nucleosome | | Descriptor: | Chromatin modification-related protein EAF3, DNA (176-MER), Histone H2A, ... | | Authors: | Markert, J.W, Vos, S.M, Farnung, L. | | Deposit date: | 2023-08-03 | | Release date: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure of the complete Saccharomyces cerevisiae Rpd3S-nucleosome complex.
Nat Commun, 14, 2023
|
|
8UW1
 
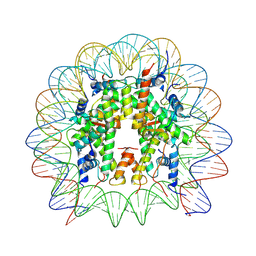 | | Cryo-EM structure of DNMT3A1 UDR in complex with H2AK119Ub-nucleosome | | Descriptor: | DNA (146-MER), DNA (cytosine-5)-methyltransferase 3A, Histone H2A, ... | | Authors: | Gretarsson, K, Abini-Agbomson, S, Armache, K.-J, Lu, C. | | Deposit date: | 2023-11-05 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Cancer-associated DNA hypermethylation of Polycomb targets requires DNMT3A dual recognition of histone H2AK119 ubiquitination and the nucleosome acidic patch.
Sci Adv, 10, 2024
|
|
8SIY
 
 | |
8HY0
 
 | |
8HXY
 
 | |
8THU
 
 | |
8U5H
 
 | |
8V25
 
 | |
8V26
 
 | |
8V27
 
 | |
8V28
 
 | |
8T9H
 
 | |
8HXZ
 
 | | Cryo-EM structure of Eaf3 CHD in complex with nucleosome | | Descriptor: | Chromatin modification-related protein EAF3, DNA (352-MER), Histone H2A, ... | | Authors: | Cui, H, Wang, H. | | Deposit date: | 2023-01-05 | | Release date: | 2023-09-27 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of histone deacetylase complex Rpd3S bound to nucleosome.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8IHM
 
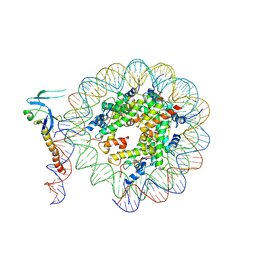 | | Eaf3 CHD domain bound to the nucleosome | | Descriptor: | Chromatin modification-related protein EAF3, DNA (164-MER), DNA (165-MER), ... | | Authors: | Zhang, Y, Gang, C. | | Deposit date: | 2023-02-23 | | Release date: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Structural basis for nucleosome binding and catalysis by the yeast Rpd3S/HDAC holoenzyme.
Cell Res., 33, 2023
|
|
8IHT
 
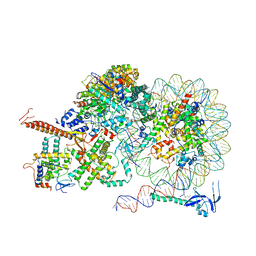 | | Rpd3S bound to the nucleosome | | Descriptor: | CALCIUM ION, Chromatin modification-related protein EAF3, DNA (164-MER), ... | | Authors: | Zhang, Y, Gang, C. | | Deposit date: | 2023-02-23 | | Release date: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Structural basis for nucleosome binding and catalysis by the yeast Rpd3S/HDAC holoenzyme.
Cell Res., 33, 2023
|
|
8JHO
 
 | |
