5MFC
 
 | | Designed armadillo repeat protein YIIIM5AII in complex with (KR)4-GFP | | Descriptor: | (KR)4-Green fluorescent protein,Green fluorescent protein, ACETATE ION, YIIIM5AII | | Authors: | Hansen, S, Kiefer, J, Madhurantakam, C, Mittl, P, Plueckthun, A. | | Deposit date: | 2016-11-18 | | Release date: | 2017-07-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of designed armadillo repeat proteins binding to peptides fused to globular domains.
Protein Sci., 26, 2017
|
|
5VMQ
 
 | |
5NGW
 
 | | Glycoside hydrolase-like protein | | Descriptor: | BROMIDE ION, OpGH99A | | Authors: | Robb, C.S, Hehemann, J.-H. | | Deposit date: | 2017-03-20 | | Release date: | 2018-01-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of a marine glycoside hydrolase family 99-related protein lacking catalytic machinery.
Protein Sci., 26, 2017
|
|
5VES
 
 | | The 2.4A crystal structure of OmpA domain of OmpA from Salmonella enterica subsp. enterica serovar Typhimurium str. 14028S | | Descriptor: | Outer membrane protein A, SULFATE ION | | Authors: | Tan, K, Wu, R, Jedrzejczak, R, Adkins, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Program for the Characterization of Secreted Effector Proteins (PCSEP) | | Deposit date: | 2017-04-05 | | Release date: | 2017-04-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Insights into PG-binding, conformational change, and dimerization of the OmpA C-terminal domains from Salmonella enterica serovar Typhimurium and Borrelia burgdorferi.
Protein Sci., 26, 2017
|
|
5VNQ
 
 | | Neutron crystallographic structure of perdeuterated T4 lysozyme cysteine-free pseudo-wild type at cryogenic temperature | | Descriptor: | CHLORIDE ION, Endolysin | | Authors: | Li, L, Shukla, S, Meilleur, F, Standaert, R.F, Pierce, J, Myles, D.A.A, Cuneo, M.J. | | Deposit date: | 2017-05-01 | | Release date: | 2017-07-26 | | Last modified: | 2023-10-04 | | Method: | NEUTRON DIFFRACTION (2.2 Å) | | Cite: | Neutron crystallographic studies of T4 lysozyme at cryogenic temperature.
Protein Sci., 26, 2017
|
|
5VMD
 
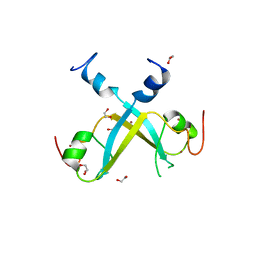 | | Crystal structure of UBR-box from UBR6 in a domain-swapping conformation | | Descriptor: | 1,2-ETHANEDIOL, F-box only protein 11, ZINC ION | | Authors: | Munoz-Escobar, J, Kozlov, G, Gehring, K. | | Deposit date: | 2017-04-27 | | Release date: | 2017-07-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Crystal structure of the UBR-box from UBR6/FBXO11 reveals domain swapping mediated by zinc binding.
Protein Sci., 26, 2017
|
|
5VSI
 
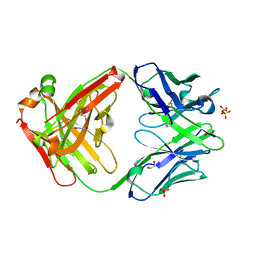 | | CH1/Ckappa Fab mutant 15.1 | | Descriptor: | 1,2-ETHANEDIOL, CH1/Ckappa Fab heavy chain, CH1/Ckappa Fab light chain, ... | | Authors: | Hendle, J. | | Deposit date: | 2017-05-11 | | Release date: | 2017-08-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Computational design of a specific heavy chain/ kappa light chain interface for expressing fully IgG bispecific antibodies.
Protein Sci., 26, 2017
|
|
5MFD
 
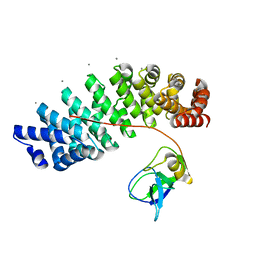 | | Designed armadillo repeat protein YIIIM''6AII in complex with pD_(KR)5 | | Descriptor: | CALCIUM ION, Capsid decoration protein,pD_(KR)5, YIIIM''6AII | | Authors: | Hansen, S, Kiefer, J, Madhurantakam, C, Mittl, P, Plueckthun, A. | | Deposit date: | 2016-11-18 | | Release date: | 2017-07-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of designed armadillo repeat proteins binding to peptides fused to globular domains.
Protein Sci., 26, 2017
|
|
5VNR
 
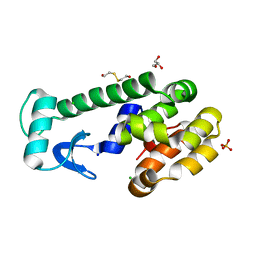 | | X-ray structure of perdeuterated T4 lysozyme cysteine-free pseudo-wild type at cryogenic temperature | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, Endolysin, ... | | Authors: | Li, L, Shukla, S, Meilleur, F, Standaert, R.F, Pierce, J, Myles, D.A.A, Cuneo, M.J. | | Deposit date: | 2017-05-01 | | Release date: | 2017-07-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.631 Å) | | Cite: | Neutron crystallographic studies of T4 lysozyme at cryogenic temperature.
Protein Sci., 26, 2017
|
|
5W71
 
 | | X-ray structure of BtrR from Bacillus circulans in the presence of the 2-DOS external aldimine | | Descriptor: | CHLORIDE ION, L-glutamine:2-deoxy-scyllo-inosose aminotransferase, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Zachman-Brockmeyer, T.R, Thoden, J.B, Holden, H.M. | | Deposit date: | 2017-06-19 | | Release date: | 2017-07-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of RbmB from Streptomyces ribosidificus, an aminotransferase involved in the biosynthesis of ribostamycin.
Protein Sci., 26, 2017
|
|
5UTV
 
 | |
5VR9
 
 | | CH1/Ckappa Fab based on Matuzumab | | Descriptor: | CH1/Ckappa Fab heavy chain, CH1/Ckappa Fab light chain | | Authors: | Hendle, J. | | Deposit date: | 2017-05-10 | | Release date: | 2017-08-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Computational design of a specific heavy chain/ kappa light chain interface for expressing fully IgG bispecific antibodies.
Protein Sci., 26, 2017
|
|
5VSH
 
 | | CH1/Clambda Fab based on Pertuzumab | | Descriptor: | CH1/Clambda Fab heavy chain, CH1/Clambda Fab light chain, SULFATE ION | | Authors: | Hendle, J. | | Deposit date: | 2017-05-11 | | Release date: | 2017-08-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Computational design of a specific heavy chain/ kappa light chain interface for expressing fully IgG bispecific antibodies.
Protein Sci., 26, 2017
|
|
5W70
 
 | | X-ray Structure of RbmB from Streptomyces ribosidificus | | Descriptor: | 1,2-ETHANEDIOL, L-glutamine:2-deoxy-scyllo-inosose aminotransferase, [4-({[(1R,2S,3S,4R,5S)-5-amino-2,3,4-trihydroxycyclohexyl]amino}methyl)-5-hydroxy-6-methylpyridin-3-yl]methyl dihydrogen phosphate | | Authors: | Zachman-Brockmeyer, T.R, Thoden, J.B, Holden, H.M. | | Deposit date: | 2017-06-19 | | Release date: | 2017-07-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of RbmB from Streptomyces ribosidificus, an aminotransferase involved in the biosynthesis of ribostamycin.
Protein Sci., 26, 2017
|
|
7KQE
 
 | |
7Z2C
 
 | | P. falciparum kinesin-8B motor domain in no nucleotide bound to tubulin dimer | | Descriptor: | Detyrosinated tubulin alpha-1B chain, GUANOSINE-5'-TRIPHOSPHATE, Kinesin-like protein, ... | | Authors: | Liu, T, Shilliday, F, Cook, A.D, Moores, C.A. | | Deposit date: | 2022-02-26 | | Release date: | 2022-10-19 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Mechanochemical tuning of a kinesin motor essential for malaria parasite transmission.
Nat Commun, 13, 2022
|
|
7Z2A
 
 | | P. berghei kinesin-8B motor domain in no nucleotide state bound to tubulin dimer | | Descriptor: | Detyrosinated tubulin alpha-1B chain, GUANOSINE-5'-TRIPHOSPHATE, Kinesin-8, ... | | Authors: | Liu, T, Shilliday, F, Cook, A.D, Moores, C.A. | | Deposit date: | 2022-02-26 | | Release date: | 2022-10-19 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Mechanochemical tuning of a kinesin motor essential for malaria parasite transmission.
Nat Commun, 13, 2022
|
|
7Z2B
 
 | | P. berghei kinesin-8B motor domain in AMPPNP state bound to tubulin dimer | | Descriptor: | Detyrosinated tubulin alpha-1B chain, GUANOSINE-5'-TRIPHOSPHATE, Kinesin-8, ... | | Authors: | Liu, T, Shilliday, F, Cook, A.D, Moores, C.A. | | Deposit date: | 2022-02-26 | | Release date: | 2022-11-16 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Mechanochemical tuning of a kinesin motor essential for malaria parasite transmission.
Nat Commun, 13, 2022
|
|
7RU6
 
 | |
7RUG
 
 | | Human SERINC3-DeltaICL4 | | Descriptor: | Serine incorporator 3, SiA | | Authors: | Purdy, M.D, Leonhardt, S.A, Yeager, M. | | Deposit date: | 2021-08-17 | | Release date: | 2022-08-24 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Antiviral HIV-1 SERINC restriction factors disrupt virus membrane asymmetry.
Nat Commun, 14, 2023
|
|
8RJC
 
 | |
8RJB
 
 | |
8RJD
 
 | |
6ZHX
 
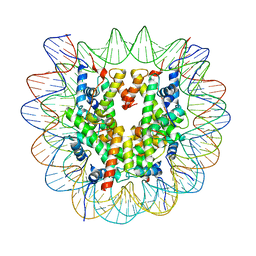 | | Cryo-EM structure of the regulatory linker of ALC1 bound to the nucleosome's acidic patch: nucleosome class. | | Descriptor: | Chromodomain-helicase-DNA-binding protein 1-like, DNA (145-MER) Widom 601 sequence, Histone H2A type 1, ... | | Authors: | Bacic, L, Gaullier, G, Croll, T.I, Deindl, S. | | Deposit date: | 2020-06-24 | | Release date: | 2020-12-23 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Mechanistic Insights into Regulation of the ALC1 Remodeler by the Nucleosome Acidic Patch.
Cell Rep, 33, 2020
|
|
6ZHY
 
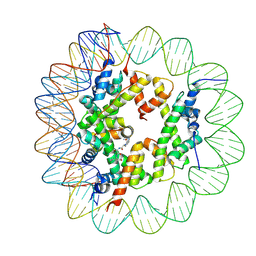 | | Cryo-EM structure of the regulatory linker of ALC1 bound to the nucleosome's acidic patch: hexasome class. | | Descriptor: | Chromodomain-helicase-DNA-binding protein 1-like, DNA (110-MER) Widom 601 sequence, Histone H2A type 1, ... | | Authors: | Bacic, L, Gaullier, G, Deindl, S. | | Deposit date: | 2020-06-24 | | Release date: | 2020-12-23 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Mechanistic Insights into Regulation of the ALC1 Remodeler by the Nucleosome Acidic Patch.
Cell Rep, 33, 2020
|
|
