1R7W
 
 | | NMR STRUCTURE OF THE R(GGAGGACAUCCCUCACGGGUGACCGUGGUCCUCC), DOMAIN IV STEM-LOOP B OF ENTEROVIRAL IRES WITH AUCCCU BULGE | | Descriptor: | 34-MER | | Authors: | Du, Z, Ulyanov, N.B, Yu, J, James, T.L. | | Deposit date: | 2003-10-22 | | Release date: | 2004-05-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR Structures of Loop B RNAs from the Stem-Loop IV Domain of the Enterovirus Internal Ribosome Entry Site: A Single C to U Substitution Drastically Changes the Shape and Flexibility of RNA(,).
Biochemistry, 43, 2004
|
|
1R7X
 
 | | Glycosyltransferase B in complex with 3-amino-acceptor analog inhibitor | | Descriptor: | Glycoprotein-fucosylgalactoside alpha-galactosyltransferase, MERCURY (II) ION, alpha-L-fucopyranose-(1-2)-hexyl 3-amino-3-deoxy-beta-D-galactopyranoside | | Authors: | Nguyen, H.P, Seto, N.O.L, Cai, Y, Leinala, E.K, Borisova, S.N, Palcic, M.M, Evans, S.V. | | Deposit date: | 2003-10-22 | | Release date: | 2004-02-10 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | The influence of an intramolecular hydrogen bond in differential recognition of inhibitory acceptor analogs by human ABO(H) blood group A and B glycosyltransferases
J.Biol.Chem., 278, 2003
|
|
1R7Y
 
 | | Glycosyltransferase A in complex with 3-amino-acceptor analog inhibitor and uridine diphosphate | | Descriptor: | Glycoprotein-fucosylgalactoside alpha-N-acetylgalactosaminyltransferase, MANGANESE (II) ION, MERCURY (II) ION, ... | | Authors: | Nguyen, H.P, Seto, N.O.L, Cai, Y, Leinala, E.K, Borisova, S.N, Palcic, M.M, Evans, S.V. | | Deposit date: | 2003-10-22 | | Release date: | 2004-02-10 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The influence of an intramolecular hydrogen bond in differential recognition of inhibitory acceptor analogs by human ABO(H) blood group A and B glycosyltransferases
J.Biol.Chem., 278, 2003
|
|
1R7Z
 
 | | NMR STRUCTURE OF THE R(GGAGGACAUUCCUCACGGGUGACCGUGGUCCUCC), DOMAIN IV STEM-LOOP B OF ENTEROVIRAL IRES WITH AUUCCU BULGE | | Descriptor: | 34-MER | | Authors: | Du, Z, Ulyanov, N.B, Yu, J, James, T.L. | | Deposit date: | 2003-10-22 | | Release date: | 2004-05-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR Structures of Loop B RNAs from the Stem-Loop IV Domain of the Enterovirus Internal Ribosome Entry Site: A Single C to U Substitution Drastically Changes the Shape and Flexibility of RNA(,).
Biochemistry, 43, 2004
|
|
1R80
 
 | | Glycosyltransferase B in complex with 3-amino-acceptor analog inhibitor and uridine diphosphate | | Descriptor: | Glycoprotein-fucosylgalactoside alpha-galactosyltransferase, MANGANESE (II) ION, MERCURY (II) ION, ... | | Authors: | Nguyen, H.P, Seto, N.O.L, Cai, Y, Leinala, E.K, Borisova, S.N, Palcic, M.M, Evans, S.V. | | Deposit date: | 2003-10-22 | | Release date: | 2004-02-10 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The influence of an intramolecular hydrogen bond in differential recognition of inhibitory acceptor analogs by human ABO(H) blood group A and B glycosyltransferases
J.Biol.Chem., 278, 2003
|
|
1R81
 
 | | Glycosyltransferase A in complex with 3-amino-acceptor analog inhibitor and uridine diphosphate-N-acetyl-galactose | | Descriptor: | Glycoprotein-fucosylgalactoside alpha-N-acetylgalactosaminyltransferase, MERCURY (II) ION, URIDINE-DIPHOSPHATE-N-ACETYLGALACTOSAMINE, ... | | Authors: | Nguyen, H.P, Seto, N.O.L, Cai, Y, Leinala, E.K, Borisova, S.N, Palcic, M.M, Evans, S.V. | | Deposit date: | 2003-10-22 | | Release date: | 2004-02-10 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The influence of an intramolecular hydrogen bond in differential recognition of inhibitory acceptor analogs by human ABO(H) blood group A and B glycosyltransferases
J.Biol.Chem., 278, 2003
|
|
1R82
 
 | | Glycosyltransferase B in complex with 3-amino-acceptor analog inhibitor, and uridine diphosphate-galactose | | Descriptor: | GALACTOSE-URIDINE-5'-DIPHOSPHATE, Glycoprotein-fucosylgalactoside alpha-galactosyltransferase, MERCURY (II) ION, ... | | Authors: | Nguyen, H.P, Seto, N.O.L, Cai, Y, Leinala, E.K, Borisova, S.N, Palcic, M.M, Evans, S.V. | | Deposit date: | 2003-10-22 | | Release date: | 2004-02-10 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The influence of an intramolecular hydrogen bond in differential recognition of inhibitory acceptor analogs by human ABO(H) blood group A and B glycosyltransferases
J.Biol.Chem., 278, 2003
|
|
1R84
 
 | | NMR structure of the 13-cis-15-syn retinal in dark_adapted bacteriorhodopsin | | Descriptor: | Bacteriorhodopsin, RETINAL | | Authors: | Patzelt, H, Simon, B, Ter Laak, A, Kessler, B, Kuhne, R, Schmieder, P, Oesterhaelt, D, Oschkinat, H. | | Deposit date: | 2003-10-23 | | Release date: | 2003-11-11 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | The structures of the active center in dark-adapted bacteriorhodopsin by solution-state NMR spectroscopy
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1R85
 
 | | Crystal structure of the extracellular xylanase from Geobacillus stearothermophilus T-6 (XT6): The WT enzyme (monoclinic form) at 1.45A resolution | | Descriptor: | CHLORIDE ION, Endo-1,4-beta-xylanase, GLYCEROL, ... | | Authors: | Bar, M, Golan, G, Nechama, M, Zolotnitsky, G, Shoham, Y, Shoham, G. | | Deposit date: | 2003-10-23 | | Release date: | 2004-07-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Mapping glycoside hydrolase substrate subsites by isothermal titration calorimetry.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
1R86
 
 | | Crystal structure of the extracellular xylanase from Geobacillus stearothermophilus T-6 (XT6, monoclinic form): The E159A/E265A mutant at 1.8A resolution | | Descriptor: | CHLORIDE ION, Endo-1,4-beta-xylanase, SULFATE ION, ... | | Authors: | Bar, M, Golan, G, Zolotnitsky, G, Shoham, Y, Shoham, G. | | Deposit date: | 2003-10-23 | | Release date: | 2005-07-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the extracellular xylanase from Geobacillus stearothermophilus T-6 (XT6, monoclinic
form): The E159A/E265A mutant at 1.8A resolution
To be Published
|
|
1R87
 
 | | Crystal structure of the extracellular xylanase from Geobacillus stearothermophilus T-6 (XT6, monoclinic form): The complex of the WT enzyme with xylopentaose at 1.67A resolution | | Descriptor: | CHLORIDE ION, Endo-1,4-beta-xylanase, SULFATE ION, ... | | Authors: | Bar, M, Golan, G, Zolotnitsky, G, Shoham, Y, Shoham, G. | | Deposit date: | 2003-10-23 | | Release date: | 2004-07-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Mapping glycoside hydrolase substrate subsites by isothermal titration calorimetry.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
1R88
 
 | | The crystal structure of Mycobacterium tuberculosis MPT51 (FbpC1) | | Descriptor: | MPT51/MPB51 antigen | | Authors: | Wilson, R.A, Maughan, W.N, Kremer, L, Besra, G.S, Futterer, K. | | Deposit date: | 2003-10-23 | | Release date: | 2003-12-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | The structure of Mycobacterium tuberculosis MPT51 (FbpC1) defines a new family of non-catalytic alpha/beta hydrolases.
J.Mol.Biol., 335, 2004
|
|
1R89
 
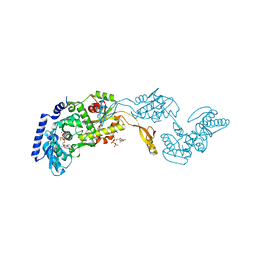 | | Crystal Structures of an Archaeal Class I CCA-Adding Enzyme and Its Nucleotide Complexes | | Descriptor: | CHLORIDE ION, CYTIDINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Xiong, Y, Li, F, Wang, J, Weiner, A.M, Steitz, T.A. | | Deposit date: | 2003-10-23 | | Release date: | 2003-12-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of an archaeal class I CCA-adding enzyme and its nucleotide complexes
Mol.Cell, 12, 2003
|
|
1R8A
 
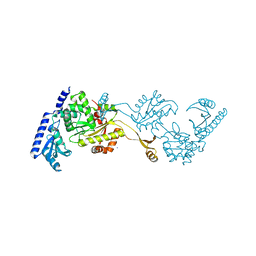 | | Crystal Structures of an Archaeal Class I CCA-Adding Enzyme and Its Nucleotide Complexes | | Descriptor: | MANGANESE (II) ION, SODIUM ION, tRNA nucleotidyltransferase | | Authors: | Xiong, Y, Li, F, Wang, J, Weiner, A.M, Steitz, T.A. | | Deposit date: | 2003-10-23 | | Release date: | 2003-12-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of an archaeal class I CCA-adding enzyme and its nucleotide complexes
Mol.Cell, 12, 2003
|
|
1R8B
 
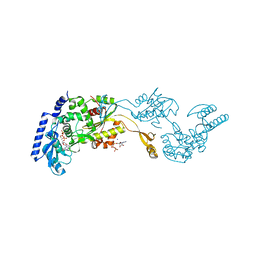 | | Crystal Structures of an Archaeal Class I CCA-Adding Enzyme and Its Nucleotide | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Xiong, Y, Li, F, Wang, J, Weiner, A.M, Steitz, T.A. | | Deposit date: | 2003-10-23 | | Release date: | 2003-12-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of an archaeal class I CCA-adding enzyme and its nucleotide complexes
Mol.Cell, 12, 2003
|
|
1R8C
 
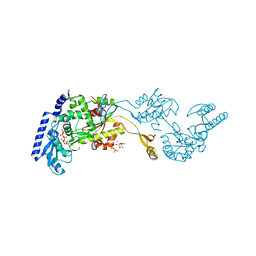 | | Crystal Structures of an Archaeal Class I CCA-Adding Enzyme and Its Nucleotide | | Descriptor: | MANGANESE (II) ION, SODIUM ION, URIDINE 5'-TRIPHOSPHATE, ... | | Authors: | Xiong, Y, Li, F, Wang, J, Weiner, A.M, Steitz, T.A. | | Deposit date: | 2003-10-23 | | Release date: | 2003-12-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of an archaeal class I CCA-adding enzyme and its nucleotide complexes
Mol.Cell, 12, 2003
|
|
1R8D
 
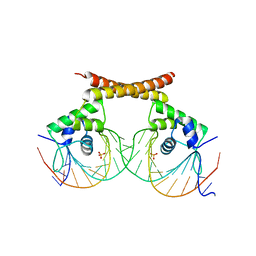 | | Crystal Structure of MtaN Bound to DNA | | Descriptor: | 26-MER, SULFATE ION, transcription activator MtaN | | Authors: | Newberry, K.J, Brennan, R.G. | | Deposit date: | 2003-10-23 | | Release date: | 2004-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structural mechanism for transcription activation by MerR family member multidrug transporter activation, N terminus.
J.Biol.Chem., 279, 2004
|
|
1R8E
 
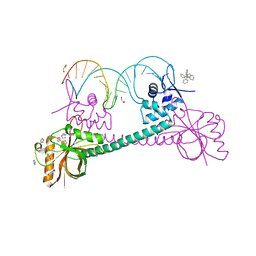 | | Crystal Structure of BmrR Bound to DNA at 2.4A Resolution | | Descriptor: | 5'-D(*GP*AP*CP*CP*CP*TP*CP*CP*CP*CP*TP*TP*AP*GP*GP*GP*GP*AP*GP*GP*GP*TP*C)-3', GLYCEROL, IMIDAZOLE, ... | | Authors: | Newberry, K.J, Brennan, R.G. | | Deposit date: | 2003-10-23 | | Release date: | 2004-08-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structural mechanism for transcription activation by MerR family member multidrug transporter activation, N terminus.
J.Biol.Chem., 279, 2004
|
|
1R8G
 
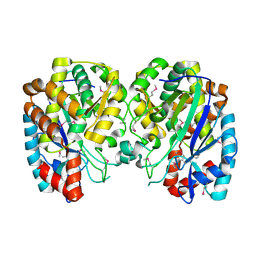 | | Structure and function of YbdK | | Descriptor: | Hypothetical protein ybdK | | Authors: | Lehmann, C, Doseeva, V, Pullalarevu, S, Krajewski, W, Howard, A, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2003-10-24 | | Release date: | 2004-08-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | YbdK is a carboxylate-amine ligase with a gamma-glutamyl:Cysteine ligase activity: crystal structure and enzymatic assays
PROTEINS: STRUCT.,FUNCT.,GENET., 56, 2004
|
|
1R8H
 
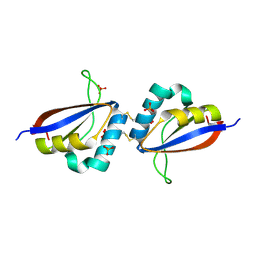 | | Comparison of the structure and DNA binding properties of the E2 proteins from an oncogenic and a non-oncogenic human papillomavirus | | Descriptor: | PHOSPHATE ION, Regulatory protein E2 | | Authors: | Dell, G, Wilkinson, K.W, Tranter, R, Parish, J, Brady, R.L, Gaston, K. | | Deposit date: | 2003-10-24 | | Release date: | 2003-12-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Comparison of the structure and DNA-binding properties of the E2 proteins from an oncogenic and a non-oncogenic human papillomavirus.
J.Mol.Biol., 334, 2003
|
|
1R8I
 
 | | Crystal structure of TraC | | Descriptor: | TraC | | Authors: | Yeo, H.-J, Yuan, Q, Beck, M.R, Baron, C, Waksman, G. | | Deposit date: | 2003-10-24 | | Release date: | 2004-01-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and functional characterization of the VirB5 protein from the type IV secretion system encoded by the conjugative plasmid pKM101
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1R8J
 
 | |
1R8K
 
 | | PDXA PROTEIN; NAD-DEPENDENT DEHYDROGENASE/CARBOXYLASE; SUBUNIT OF PYRIDOXINE PHOSPHATE BIOSYNTHETIC PROTEIN PDXJ-PDXA [SALMONELLA TYPHIMURIUM] | | Descriptor: | 4-hydroxythreonine-4-phosphate dehydrogenase 1, CHLORIDE ION, COBALT (II) ION, ... | | Authors: | Osipiuk, J, Quartey, P, Moy, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-10-27 | | Release date: | 2003-11-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of NAD-dependent dehydrogenase/carboxylase of Salmonella typhimurium
to be published
|
|
1R8L
 
 | | The structure of endo-beta-1,4-galactanase from Bacillus licheniformis | | Descriptor: | CALCIUM ION, endo-beta-1,4-galactanase | | Authors: | Ryttersgaard, C, Le Nours, J, Lo Leggio, L, Jorgensen, C.T, Christensen, L.L, Bjornvad, M, Larsen, S. | | Deposit date: | 2003-10-27 | | Release date: | 2004-11-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structure of endo-beta-1,4-galactanase from Bacillus licheniformis in complex with two oligosaccharide products
J.Mol.Biol., 341, 2004
|
|
1R8M
 
 | |
