1WCC
 
 | | Screening for fragment binding by X-ray crystallography | | Descriptor: | 2-AMINO-6-CHLOROPYRAZINE, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Cleasby, A, O'Reilly, M, Hartshorn, M.J, Murray, C.W, Tickle, I.J, Jhoti, H, Frederickson, M. | | Deposit date: | 2004-11-12 | | Release date: | 2005-01-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Fragment-Based Lead Discovery Using X-Ray Crystallography
J.Med.Chem., 48, 2005
|
|
1WCD
 
 | | Crystal structure of IBDV T1 virus-like particle reveals a missing link in icosahedral viruses evolution | | Descriptor: | MAJOR STRUCTURAL PROTEIN VP2 | | Authors: | Coulibaly, F, Chevalier, C, Gutsche, I, Pous, J, Bressanelli, S, Navaza, J, Delmas, B, Rey, F.A. | | Deposit date: | 2004-11-12 | | Release date: | 2005-04-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Birnavirus Crystal Structure Reveals Structural Relationships Among Icosahedral Viruses.
Cell(Cambridge,Mass.), 120, 2005
|
|
1WCE
 
 | | Crystal structure of the T13 IBDV viral particle reveals a missing link in icosahedral viruses evolution | | Descriptor: | MAJOR STRUCTURAL PROTEIN VP2 | | Authors: | Coulibaly, F, Chevalier, C, Gutsche, I, Pous, J, Bressanelli, S, Navaza, J, Delmas, B, Rey, F.A. | | Deposit date: | 2004-11-12 | | Release date: | 2005-04-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (7 Å) | | Cite: | The Birnavirus Crystal Structure Reveals Structural Relationships Among Icosahedral Viruses.
Cell(Cambridge,Mass.), 120, 2005
|
|
1WCF
 
 | |
1WCG
 
 | | Aphid myrosinase | | Descriptor: | GLYCEROL, THIOGLUCOSIDASE | | Authors: | Husebye, H, Arzt, S, Burmeister, W.P, Haertel, F.V, Brandt, A, Rossiter, J.T, Bones, A.M. | | Deposit date: | 2004-11-15 | | Release date: | 2005-11-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Crystal Structure at 1.1A Resolution of an Insect Myrosinase from Brevicoryne Brassicae Shows its Close Relationship to Beta-Glucosidases.
Insect Biochem.Mol.Biol., 35, 2005
|
|
1WCH
 
 | | Crystal structure of PTPL1 human tyrosine phosphatase mutated in colorectal cancer - evidence for a second phosphotyrosine substrate recognition pocket | | Descriptor: | PHOSPHATE ION, PROTEIN TYROSINE PHOSPHATASE, NON-RECEPTOR TYPE 13 | | Authors: | Villa, F, Deak, M, Bloomberg, G.B, Alessi, D.R, Van Aalten, D.M.F. | | Deposit date: | 2004-11-16 | | Release date: | 2004-12-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of Ptpl1/Fap-1 Human Tyrosine Phosphatase Mutated in Colorectal Cancer: Eveidence for a Second Phosphotyrosine Substrate Recognition Pocket
J.Biol.Chem., 280, 2005
|
|
1WCI
 
 | | Reactivity modulation of human branched-chain alpha-ketoacid dehydrogenase by an internal molecular switch | | Descriptor: | 2-OXOISOVALERATE DEHYDROGENASE ALPHA SUBUNIT, 2-OXOISOVALERATE DEHYDROGENASE BETA SUBUNIT, C2-1-HYDROXY-3-METHYL-BUTYL-THIAMIN, ... | | Authors: | Machius, M, Wynn, R.M, Chuang, J.L, Tomchick, D.R, Brautigam, C.A, Chuang, D.T. | | Deposit date: | 2004-11-16 | | Release date: | 2006-02-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | A Versatile Conformational Switch Regulates Reactivity in Human Branched-Chain Alpha-Ketoacid Dehydrogenase.
Structure, 14, 2006
|
|
1WCJ
 
 | | Conserved Hypothetical Protein TM0487 from Thermotoga maritima | | Descriptor: | HYPOTHETICAL PROTEIN TM0487 | | Authors: | Almeida, M.S, Peti, W, Herrmann, T, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2004-11-17 | | Release date: | 2004-12-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Conserved Hypothetical Protein Tm0487 from Thermotoga Maritima: Implications for 216 Homologous Duf59 Proteins.
Protein Sci., 14, 2005
|
|
1WCK
 
 | | Crystal structure of the C-terminal domain of BclA, the major antigen of the exosporium of the Bacillus anthracis spore. | | Descriptor: | BCLA PROTEIN, CACODYLATE ION | | Authors: | Rety, S, Salamitou, S, Augusto, L.A, Chaby, R, Lehegarat, F, Lewit-Bentley, A. | | Deposit date: | 2004-11-17 | | Release date: | 2005-10-25 | | Last modified: | 2019-03-06 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | The Crystal Structure of the Bacillus Anthracis Spore Surface Protein Bcla Shows Remarkable Similarity to Mammalian Proteins.
J.Biol.Chem., 280, 2005
|
|
1WCL
 
 | |
1WCM
 
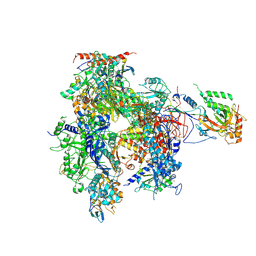 | | Complete 12-Subunit RNA Polymerase II at 3.8 Angstrom | | Descriptor: | DNA-DIRECTED RNA POLYMERASE II 13.6 KDA POLYPEPTIDE, DNA-DIRECTED RNA POLYMERASE II 14.2 KDA POLYPEPTIDE, DNA-DIRECTED RNA POLYMERASE II 19 KD POLYPEPTIDE, ... | | Authors: | Armache, K.-J, Mitterweger, S, Meinhart, A, Cramer, P. | | Deposit date: | 2004-11-17 | | Release date: | 2004-12-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structures of Complete RNA Polymerase II and its Subcomplex,Rpb4/7
J.Biol.Chem., 280, 2005
|
|
1WCN
 
 | |
1WCO
 
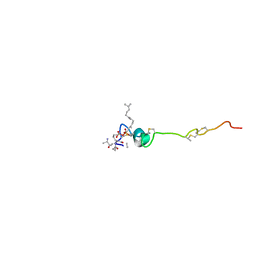 | | The solution structure of the nisin-lipid II complex | | Descriptor: | (2E,6E)-12-fluoro-11-(fluoromethyl)-3,7-dimethyldodeca-2,6,10-trien-1-yl trihydrogen diphosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-alpha-muramic acid, ALA-FGA-LYS-DAL-DAL PEPTIDE, ... | | Authors: | Hsu, S.-T.D, Breukink, E, Tischenko, E, Lutters, M.A.G, de Kruijff, B, Kaptein, R, Bonvin, A.M.J.J, van Nuland, N.A.J. | | Deposit date: | 2004-11-19 | | Release date: | 2005-03-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The nisin-lipid II complex reveals a pyrophosphate cage that provides a blueprint for novel antibiotics.
Nat. Struct. Mol. Biol., 11, 2004
|
|
1WCQ
 
 | | Mutagenesis of the Nucleophilic Tyrosine in a Bacterial Sialidase to Phenylalanine. | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, GLYCEROL, SIALIDASE, ... | | Authors: | Newstead, S, Watson, J.N, Bennet, A.J, Taylor, G. | | Deposit date: | 2004-11-19 | | Release date: | 2005-10-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Two Nucleophilic Mutants of the Micromonospora Viridifaciens Sialidase Operate with Retention of Configuration by Two Different Mechanisms.
Chembiochem, 6, 2005
|
|
1WCR
 
 | |
1WCS
 
 | | A mutant of Trypanosoma rangeli sialidase displaying trans-sialidase activity | | Descriptor: | SIALIDASE | | Authors: | Paris, G, Ratier, L, Amaya, M.F, Nguyen, T, Alzari, P.M, Frasch, C. | | Deposit date: | 2004-11-21 | | Release date: | 2004-12-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A Sialidase Mutant Displaying Trans-Sialidase Activity
J.Mol.Biol., 345, 2005
|
|
1WCT
 
 | | A NOVEL CONOTOXIN FROM CONUS TEXTILE WITH UNUSUAL POST-TRANSLATIONAL MODIFICATIONS REDUCES PRESYNAPTIC CALCIUM INFLUX, NMR, 1 STRUCTURE, GLYCOSYLATED PROTEIN | | Descriptor: | OMEGAC-TXIX, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose | | Authors: | Rigby, A.C, Hambe, B, Czerwiec, E, Baleja, J.D, Furie, B.C, Furie, B, Stenflo, J. | | Deposit date: | 1998-12-18 | | Release date: | 1999-06-08 | | Last modified: | 2020-07-29 | | Method: | SOLUTION NMR | | Cite: | A conotoxin from Conus textile with unusual posttranslational modifications reduces presynaptic Ca2+ influx.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1WCU
 
 | | CBM29_1, A Family 29 Carbohydrate Binding Module from Piromyces equi | | Descriptor: | GLYCEROL, NON-CATALYTIC PROTEIN 1 | | Authors: | Flint, J, Bolam, D.N, Nurizzo, D, Taylor, E.J, Williamson, M.P, Walters, C, Davis, G.J, Gilbert, H.J. | | Deposit date: | 2004-11-22 | | Release date: | 2005-03-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Probing the Mechanism of Ligand Recognition in Family 29 Carbohydrate-Binding Modules
J.Biol.Chem., 280, 2005
|
|
1WCV
 
 | |
1WCW
 
 | | Crystal Structure of Uroporphyrinogen III Synthase from an Extremely Thermophilic Bacterium Thermus thermophilus HB8 (Wild type, Native, Form-1 crystal) | | Descriptor: | GLYCEROL, Uroporphyrinogen III synthase | | Authors: | Mizohata, E, Matsuura, T, Sakai, H, Murayama, K, Terada, T, Shirouzu, M, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-06 | | Release date: | 2005-05-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal Structure of Uroporphyrinogen III Synthase from Thermus thermophilus HB8
To be Published
|
|
1WCX
 
 | | Crystal Structure of Mutant Uroporphyrinogen III Synthase from an Extremely Thermophilic Bacterium Thermus thermophilus HB8 (L75M/I193M/L248M, SeMet derivative, Form-1 crystal) | | Descriptor: | GLYCEROL, Uroporphyrinogen III Synthase | | Authors: | Mizohata, E, Matsuura, T, Murayama, K, Sakai, H, Terada, T, Shirouzu, M, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-06 | | Release date: | 2005-05-06 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Uroporphyrinogen III Synthase from Thermus thermophilus HB8
To be Published
|
|
1WCY
 
 | | Crystal Structure Of Human Dipeptidyl Peptidase IV (DPPIV) Complex With Diprotin A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase IV, ... | | Authors: | Hiramatsu, H, Yamamoto, A, Kyono, K, Higashiyama, Y, Fukushima, C, Shima, H, Sugiyama, S, Inaka, K, Shimizu, R. | | Deposit date: | 2004-05-07 | | Release date: | 2005-05-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of human dipeptidyl peptidase IV (DPPIV) complex with diprotin A
Biol.Chem., 385, 2004
|
|
1WCZ
 
 | | Crystal structure of an alkaline form of v8 protease from Staphylococcus aureus | | Descriptor: | Glutamyl endopeptidase, ZINC ION | | Authors: | Yamada, K, Ohta, M, Hasegawa, T, Torii, K, Murakami, M, Kouyama, K. | | Deposit date: | 2004-05-10 | | Release date: | 2004-06-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of an alkaline form of v8 protease from Staphylococcus aureus
To be Published
|
|
1WD0
 
 | | Crystal structures of the hyperthermophilic chromosomal protein Sac7d in complex with DNA decamers | | Descriptor: | 5'-D(*CP*CP*TP*AP*TP*AP*TP*AP*GP*G)-3', DNA-binding proteins 7a/7b/7d | | Authors: | Ko, T.-P, Chu, H.-M, Chen, C.-Y, Chou, C.-C, Wang, A.H.-J. | | Deposit date: | 2004-05-10 | | Release date: | 2004-08-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of the hyperthermophilic chromosomal protein Sac7d in complex with DNA decamers.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1WD1
 
 | | Crystal structures of the hyperthermophilic chromosomal protein Sac7d in complex with DNA decamers | | Descriptor: | 5'-D(*CP*CP*TP*AP*CP*GP*TP*AP*GP*G)-3', DNA-binding proteins 7a/7b/7d | | Authors: | Ko, T.-P, Chu, H.-M, Chen, C.-Y, Chou, C.-C, Wang, A.H.-J. | | Deposit date: | 2004-05-10 | | Release date: | 2004-08-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of the hyperthermophilic chromosomal protein Sac7d in complex with DNA decamers.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
