2K6P
 
 | | Solution Structure of hypothetical protein, HP1423 | | Descriptor: | Uncharacterized protein HP_1423 | | Authors: | Kim, J, Park, S, Lee, K, Son, W, Sohn, N, Lee, B. | | Deposit date: | 2008-07-15 | | Release date: | 2009-06-16 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of hypothetical protein HP1423 (Y1423_HELPY) reveals the presence of alphaL motif related to RNA binding
Proteins, 75, 2009
|
|
2CQJ
 
 | | Solution structure of the S4 domain of U3 small nucleolar ribonucleoprotein protein IMP3 homolog | | Descriptor: | U3 small nucleolar ribonucleoprotein protein IMP3 homolog | | Authors: | Suzuki, S, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-20 | | Release date: | 2005-11-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the S4 domain of U3 small nucleolar ribonucleoprotein protein IMP3 homolog
To be Published
|
|
3BBU
 
 | |
1DM9
 
 | | HEAT SHOCK PROTEIN 15 KD | | Descriptor: | HYPOTHETICAL 15.5 KD PROTEIN IN MRCA-PCKA INTERGENIC REGION, SULFATE ION | | Authors: | Staker, B.L, Korber, P, Bardwell, J.C.A, Saper, M.A. | | Deposit date: | 1999-12-14 | | Release date: | 2000-02-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Hsp15 reveals a novel RNA-binding motif.
EMBO J., 19, 2000
|
|
5Z81
 
 | |
3HP7
 
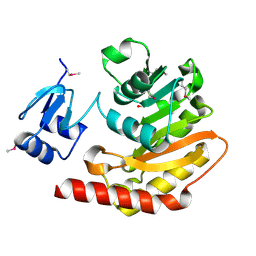 | | Putative hemolysin from Streptococcus thermophilus. | | Descriptor: | ETHANOL, Hemolysin, putative | | Authors: | Osipiuk, J, Hatzos, C, Cobb, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-06-03 | | Release date: | 2009-06-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | X-ray crystal structure of putative hemolysin from Streptococcus thermophilus.
To be Published
|
|
3DH3
 
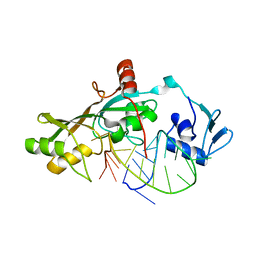 | | Crystal Structure of RluF in complex with a 22 nucleotide RNA substrate | | Descriptor: | Ribosomal large subunit pseudouridine synthase F, stem loop fragment of E. Coli 23S RNA | | Authors: | Alian, A, DeGiovanni, A, Stroud, R.M, Finer-Moore, J.S. | | Deposit date: | 2008-06-16 | | Release date: | 2009-04-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of an RluF-RNA complex: a base-pair rearrangement is the key to selectivity of RluF for U2604 of the ribosome.
J.Mol.Biol., 388, 2009
|
|
1QYU
 
 | |
3OTO
 
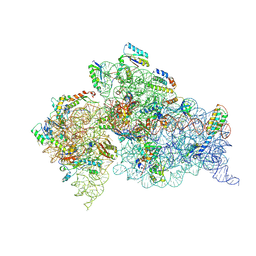 | | Crystal Structure of the 30S ribosomal subunit from a KsgA mutant of Thermus thermophilus (HB8) | | Descriptor: | 16S rRNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Demirci, H, Murphy IV, F, Belardinelli, R, Kelley, A.C, Ramakrishnan, V, Gregory, S.T, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2010-09-13 | | Release date: | 2010-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.69 Å) | | Cite: | Modification of 16S ribosomal RNA by the KsgA methyltransferase restructures the 30S subunit to optimize ribosome function.
Rna, 16, 2010
|
|
3J77
 
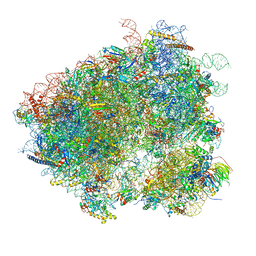 | | Structures of yeast 80S ribosome-tRNA complexes in the rotated and non-rotated conformations (Class II - rotated ribosome with 1 tRNA) | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0, ... | | Authors: | Svidritskiy, E, Brilot, A.F, Koh, C.S, Grigorieff, N, Korostelev, A.A. | | Deposit date: | 2014-05-29 | | Release date: | 2014-08-06 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Structures of Yeast 80S Ribosome-tRNA Complexes in the Rotated and Nonrotated Conformations.
Structure, 22, 2014
|
|
3JA1
 
 | | Activation of GTP Hydrolysis in mRNA-tRNA Translocation by Elongation Factor G | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Li, W, Liu, Z, Koripella, R.K, Langlois, R, Sanyal, S, Frank, J. | | Deposit date: | 2015-03-30 | | Release date: | 2015-07-01 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Activation of GTP hydrolysis in mRNA-tRNA translocation by elongation factor G.
Sci Adv, 1, 2015
|
|
6TBV
 
 | |
6TC3
 
 | |
3T1H
 
 | | Structure of the Thermus thermophilus 30S ribosomal subunit complexed with a human anti-codon stem loop (HASL) of transfer RNA lysine 3 (tRNALys3) bound to an mRNA with an AAA-codon in the A-site and Paromomycin | | Descriptor: | 16s rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Murphy, F.V, Vendeix, F.A.P, Cantara, W, Leszczynska, G, Gustilo, E.M, Sproat, B, Malkiewicz, A.A.P, Agris, P.F. | | Deposit date: | 2011-07-21 | | Release date: | 2012-01-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Human tRNA(Lys3)(UUU) Is Pre-Structured by Natural Modifications for Cognate and Wobble Codon Binding through Keto-Enol Tautomerism.
J.Mol.Biol., 416, 2012
|
|
3T1Y
 
 | | Structure of the Thermus thermophilus 30S ribosomal subunit complexed with a human anti-codon stem loop (HASL) of transfer RNA Lysine 3 (TRNALYS3) bound to an mRNA with an AAG-codon in the A-site and paromomycin | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Murphy, F.V, Vendeix, F.A.P, Cantara, W, Leszczynska, G, Gustilo, E.M, Sproat, B, Malkiewicz, A.A.P, Agris, P.F. | | Deposit date: | 2011-07-22 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Human tRNA(Lys3)(UUU) Is Pre-Structured by Natural Modifications for Cognate and Wobble Codon Binding through Keto-Enol Tautomerism.
J.Mol.Biol., 416, 2012
|
|
2E5L
 
 | | A snapshot of the 30S ribosomal subunit capturing mRNA via the Shine- Dalgarno interaction | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Kaminishi, T, Wilson, D.N, Takemoto, C, Harms, J.M, Kawazoe, M, Schluenzen, F, Hanawa-Suetsugu, K, Shirouzu, M, Fucini, P, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-12-21 | | Release date: | 2007-05-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | A snapshot of the 30S ribosomal subunit capturing mRNA via the Shine-Dalgarno interaction
Structure, 15, 2007
|
|
5OQL
 
 | | Cryo-EM structure of the 90S pre-ribosome from Chaetomium thermophilum | | Descriptor: | 35S rRNA, 40S ribosomal protein S1, 40S ribosomal protein S11-like protein, ... | | Authors: | Cheng, J, Kellner, N, Berninghausen, O, Hurt, E, Beckmann, R. | | Deposit date: | 2017-08-12 | | Release date: | 2017-10-11 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | 3.2- angstrom -resolution structure of the 90S preribosome before A1 pre-rRNA cleavage.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5ON6
 
 | | Crystal structure of haemanthamine bound to the 80S ribosome | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Pellegrino, S, Meyer, M, Yusupova, G, Yusupov, M. | | Deposit date: | 2017-08-03 | | Release date: | 2018-02-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.10000229 Å) | | Cite: | The Amaryllidaceae Alkaloid Haemanthamine Binds the Eukaryotic Ribosome to Repress Cancer Cell Growth.
Structure, 26, 2018
|
|
5OPT
 
 | | Structure of KSRP in context of Trypanosoma cruzi 40S | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, putative, ... | | Authors: | Brito Querido, J, Mancera-Martinez, E, Vicens, Q, Bochler, A, Chicher, J, Simonetti, A, Hashem, Y. | | Deposit date: | 2017-08-10 | | Release date: | 2017-11-15 | | Last modified: | 2017-12-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | The cryo-EM Structure of a Novel 40S Kinetoplastid-Specific Ribosomal Protein.
Structure, 25, 2017
|
|
8CQW
 
 | | Crystal structure of the Candida albicans 80S ribosome in complex with Hygromycin B | | Descriptor: | 18S, 25S, 3-O-acetyl-2-O-(3-O-acetyl-6-deoxy-beta-D-glucopyranosyl)-6-deoxy-1-O-{[(2R,2'S,3a'R,4''S,5''R,6'S,7a'S)-5''-methyl-4''-{[(2E)-3-phenylprop-2-enoyl]oxy}decahydrodispiro[oxirane-2,3'-[1]benzofuran-2',2''-pyran]-6'-yl]carbonyl}-beta-D-glucopyranose, ... | | Authors: | Kolosova, O, Zgadzay, Y, Yusupov, M. | | Deposit date: | 2023-03-07 | | Release date: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Drug-induced rotational movement of the ribosome is a key factor for read-through enhancement
To Be Published
|
|
8CRE
 
 | | Crystal structure of the Candida albicans 80S ribosome in complex with geneticin G418 | | Descriptor: | 18S, 25S, 3-O-acetyl-2-O-(3-O-acetyl-6-deoxy-beta-D-glucopyranosyl)-6-deoxy-1-O-{[(2R,2'S,3a'R,4''S,5''R,6'S,7a'S)-5''-methyl-4''-{[(2E)-3-phenylprop-2-enoyl]oxy}decahydrodispiro[oxirane-2,3'-[1]benzofuran-2',2''-pyran]-6'-yl]carbonyl}-beta-D-glucopyranose, ... | | Authors: | Kolosova, O, Zgadzay, Y, Yusupov, M. | | Deposit date: | 2023-03-08 | | Release date: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Drug-induced rotational movement of the ribosome is a key factor for read-through enhancement
To Be Published
|
|
8CQ7
 
 | | Crystal structure of phyllanthoside bound to the Candida albicans 80S ribosome | | Descriptor: | 18S, 25S, 3-O-acetyl-2-O-(3-O-acetyl-6-deoxy-beta-D-glucopyranosyl)-6-deoxy-1-O-{[(2R,2'S,3a'R,4''S,5''R,6'S,7a'S)-5''-methyl-4''-{[(2E)-3-phenylprop-2-enoyl]oxy}decahydrodispiro[oxirane-2,3'-[1]benzofuran-2',2''-pyran]-6'-yl]carbonyl}-beta-D-glucopyranose, ... | | Authors: | Kolosova, O, Zgadzay, Y, Yusupov, M. | | Deposit date: | 2023-03-03 | | Release date: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Drug-induced rotational movement of the ribosome is a key factor for read-through enhancement
To Be Published
|
|
2F4V
 
 | | 30S ribosome + designer antibiotic | | Descriptor: | (2R)-4-AMINO-N-{(1R,2S,3R,4R,5S)-5-AMINO-2-{2-[(2-AMINOETHYL)AMINO]ETHOXY}-4-[(2,6-DIAMINO-2,6-DIDEOXY-ALPHA-D-GLUCOPYRANOSYL)OXY]-3-HYDROXYCYCLOHEXYL}-2-HYDROXYBUTANAMIDE, (2S,4S,4AR,5AS,6S,11R,11AS,12R,12AR)-7-CHLORO-4-(DIMETHYLAMINO)-6,10,11,12-TETRAHYDROXY-1,3-DIOXO-1,2,3,4,4A,5,5A,6,11,11A,12,12A-DODECAHYDROTETRACENE-2-CARBOXAMIDE, 16S ribosomal RNA, ... | | Authors: | Murray, J.B, Meroueh, S.O, Russell, R.J, Lentzen, G, Haddad, J, Mobashery, S. | | Deposit date: | 2005-11-24 | | Release date: | 2006-07-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Interactions of designer antibiotics and the bacterial ribosome aminoacyl-tRNA site
Chem.Biol., 13, 2006
|
|
2HHH
 
 | | Crystal structure of kasugamycin bound to the 30S ribosomal subunit | | Descriptor: | (1S,2R,3S,4R,5S,6S)-2,3,4,5,6-PENTAHYDROXYCYCLOHEXYL 2-AMINO-4-{[CARBOXY(IMINO)METHYL]AMINO}-2,3,4,6-TETRADEOXY-ALPHA-D-ARABINO-HEXOPYRANOSIDE, 16S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Schluenzen, F. | | Deposit date: | 2006-06-28 | | Release date: | 2006-09-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | The antibiotic kasugamycin mimics mRNA nucleotides to destabilize tRNA binding and inhibit canonical translation initiation.
Nat.Struct.Mol.Biol., 13, 2006
|
|
2IST
 
 | |
