4A4D
 
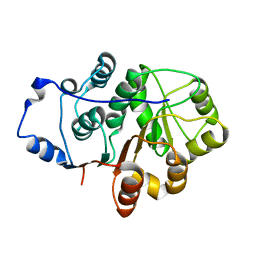 | | Crystal structure of the N-terminal domain of the Human DEAD-BOX RNA helicase DDX5 (P68) | | Descriptor: | PROBABLE ATP-DEPENDENT RNA HELICASE DDX5 | | Authors: | Dutta, S, Choi, Y.W, Kotaka, M, Fielding, B.C, Tan, Y.J. | | Deposit date: | 2011-10-11 | | Release date: | 2012-08-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Variable N-Terminal Region of Ddx5 Contains Structural Elements and Auto-Inhibits its Interaction with Ns5B of Hepatitis C Virus.
Biochem.J., 446, 2012
|
|
3PSK
 
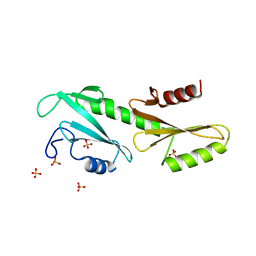 | |
1D4C
 
 | | CRYSTAL STRUCTURE OF THE UNCOMPLEXED FORM OF THE FLAVOCYTOCHROME C FUMARATE REDUCTASE OF SHEWANELLA PUTREFACIENS STRAIN MR-1 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, FLAVOCYTOCHROME C FUMARATE REDUCTASE, HEME C, ... | | Authors: | Leys, D, Tsapin, A.S, Meyer, T.E, Cusanovich, M.A, Van Beeumen, J.J. | | Deposit date: | 1999-10-03 | | Release date: | 1999-12-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure and mechanism of the flavocytochrome c fumarate reductase of Shewanella putrefaciens MR-1.
Nat.Struct.Biol., 6, 1999
|
|
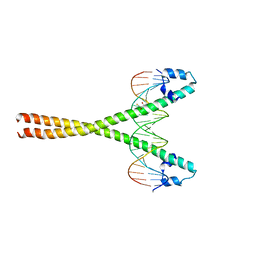
2OBH
 
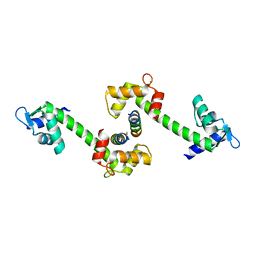 | | Centrin-XPC peptide | | Descriptor: | CALCIUM ION, Centrin-2, DNA-repair protein complementing XP-C cells | | Authors: | Charbonnier, J.B. | | Deposit date: | 2006-12-19 | | Release date: | 2007-10-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural, thermodynamic, and cellular characterization of human centrin 2 interaction with xeroderma pigmentosum group C protein.
J.Mol.Biol., 373, 2007
|
|
1W6X
 
 | | SH3 domain of p40phox, component of the NADPH oxidase | | Descriptor: | NEUTROPHIL CYTOSOL FACTOR 4 | | Authors: | Massenet, C, Chenavas, S, Cohen-Addad, C, Dagher, M.-C, Brandolin, G, Pebay-Peyroula, E, Fieschi, F. | | Deposit date: | 2004-08-24 | | Release date: | 2005-01-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Effects of P47Phox C-Terminus Phosphorylation on Binding Interactions with P40Phox and P67Phox: Structural and Functional Comparison of P40Phox P67Phox SH3 Domains
J.Biol.Chem., 280, 2005
|
|
4AUW
 
 | | CRYSTAL STRUCTURE OF THE BZIP HOMODIMERIC MAFB IN COMPLEX WITH THE C- MARE BINDING SITE | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, C-MARE BINDING SITE (5'-D(*AP*TP*AP*AP*TP*GP*CP*TP* GP*AP*CP*GP*TP*CP*AP*GP*CP*AP*AP*TP*T)-3'), MERCURY (II) ION, ... | | Authors: | Textor, L.C, Holton, S, Wilmanns, M. | | Deposit date: | 2012-05-22 | | Release date: | 2013-06-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Expression, purification, crystallization and preliminary crystallographic analysis of the mouse transcription factor MafB in complex with its DNA-recognition motif Cmare
Acta Crystallogr Sect F Struct Biol Cryst Commun., 63, 2007
|
|
3ZH8
 
 | | A novel small molecule aPKC inhibitor | | Descriptor: | (2S)-3-phenyl-N~1~-[2-(pyridin-4-yl)-5,6,7,8-tetrahydro[1]benzothieno[2,3-d]pyrimidin-4-yl]propane-1,2-diamine, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Kjaer, S, Purkiss, A.G, Kostelecky, B, Knowles, P.P, Soriano, E, Murray-Rust, J, McDonald, N.Q. | | Deposit date: | 2012-12-20 | | Release date: | 2013-02-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.739 Å) | | Cite: | Adenosine-Binding Motif Mimicry and Cellular Effects of a Thieno[2,3-D]Pyrimidine-Based Chemical Inhibitor of Atypical Protein Kinase C Isozymes.
Biochem.J., 451, 2013
|
|
1FCD
 
 | | THE STRUCTURE OF FLAVOCYTOCHROME C SULFIDE DEHYDROGENASE FROM A PURPLE PHOTOTROPHIC BACTERIUM CHROMATIUM VINOSUM AT 2.5 ANGSTROMS RESOLUTION | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, FLAVOCYTOCHROME C SULFIDE DEHYDROGENASE (CYTOCHROME SUBUNIT), FLAVOCYTOCHROME C SULFIDE DEHYDROGENASE (FLAVIN-BINDING SUBUNIT), ... | | Authors: | Chen, Z.W, Koh, M, Van Driessche, G, Van Beeumen, J.J, Bartsch, R.G, Meyer, T.E, Cusanovich, M.A, Mathews, F.S. | | Deposit date: | 1994-08-18 | | Release date: | 1994-11-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | The structure of flavocytochrome c sulfide dehydrogenase from a purple phototrophic bacterium.
Science, 266, 1994
|
|
4IWD
 
 | | Structure of dually phosphorylated c-MET receptor kinase in complex with an MK-8033 analog | | Descriptor: | 1-{5-oxo-3-[1-(piperidin-4-yl)-1H-pyrazol-4-yl]-5H-benzo[4,5]cyclohepta[1,2-b]pyridin-7-yl}-N-(pyridin-2-ylmethyl)methanesulfonamide, Hepatocyte growth factor receptor | | Authors: | Soisson, S.M, Northrup, A, Rickert, K, Patel, S, Allison, T. | | Deposit date: | 2013-01-23 | | Release date: | 2013-12-11 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Discovery of 1-[3-(1-methyl-1H-pyrazol-4-yl)-5-oxo-5H-benzo[4,5]cyclohepta[1,2-b]pyridin-7-yl]-N-(pyridin-2-ylmethyl)methanesulfonamide (MK-8033): A Specific c-Met/Ron dual kinase inhibitor with preferential affinity for the activated state of c-Met.
J.Med.Chem., 56, 2013
|
|
1YEB
 
 | |
1IRV
 
 | |
3IWZ
 
 | | The c-di-GMP Responsive Global Regulator CLP Links Cell-Cell Signaling to Virulence Gene Expression in Xanthomonas campestris | | Descriptor: | Catabolite activation-like protein | | Authors: | Chin, K.H, Tu, Z.L, Tseng, Y.H, Dow, J.M, Wang, A.H.J, Chou, S.H. | | Deposit date: | 2009-09-03 | | Release date: | 2009-12-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The cAMP receptor-like protein CLP is a novel c-di-GMP receptor linking cell-cell signaling to virulence gene expression in Xanthomonas campestris.
J.Mol.Biol., 396, 2010
|
|
2Q3X
 
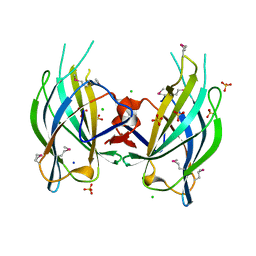 | | The RIM1alpha C2B domain | | Descriptor: | CHLORIDE ION, Regulating synaptic membrane exocytosis protein 1, SODIUM ION, ... | | Authors: | Guan, R, Dai, H, Tomchick, D.R, Machius, M, Sudhof, T.C, Rizo, J. | | Deposit date: | 2007-05-30 | | Release date: | 2007-08-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal Structure of the RIM1alpha C(2)B Domain at 1.7 A Resolution.
Biochemistry, 46, 2007
|
|
2PRE
 
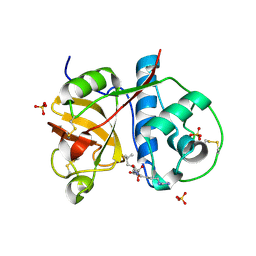 | | Crystal structure of plant cysteine protease Ervatamin-C complexed with irreversible inhibitor E-64 at 2.7 A resolution | | Descriptor: | Ervatamin-C, N-[N-[1-HYDROXYCARBOXYETHYL-CARBONYL]LEUCYLAMINO-BUTYL]-GUANIDINE, SULFATE ION | | Authors: | Ghosh, R, Chakrabarti, C, Dattagupta, J.K, Biswas, S. | | Deposit date: | 2007-05-04 | | Release date: | 2008-01-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insights into the substrate specificity and activity of ervatamins, the papain-like cysteine proteases from a tropical plant, Ervatamia coronaria.
Febs J., 275, 2008
|
|
2HJ8
 
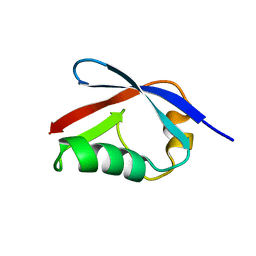 | | Solution NMR structure of the C-terminal domain of the interferon alpha-inducible ISG15 protein from Homo sapiens. Northeast Structural Genomics target HR2873B | | Descriptor: | Interferon-induced 17 kDa protein | | Authors: | Aramini, J.M, Ho, C.K, Yin, C, Cunningham, K, Janjua, H, Ma, L.-C, Xiao, R, Acton, T.B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-06-30 | | Release date: | 2006-08-01 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the C-terminal domain of the interferon alpha-inducible ISG15 protein from Homo sapiens. Northeast Structural Genomics target HR2873B.
To be Published
|
|
2HV4
 
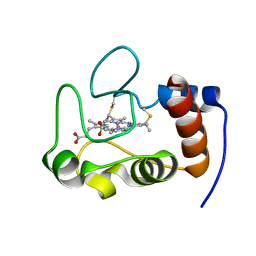 | | NMR solution structure refinement of yeast iso-1-ferrocytochrome c | | Descriptor: | Cytochrome c iso-1, HEME C | | Authors: | Assfalg, M, Bertini, I, Del Conte, R, Turano, P. | | Deposit date: | 2006-07-27 | | Release date: | 2006-09-26 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Cytochrome c and organic molecules: solution structure of the p-aminophenol adduct.
Biochemistry, 46, 2007
|
|
2FRC
 
 | | CYTOCHROME C (REDUCED) FROM EQUUS CABALLUS, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | CYTOCHROME C, HEME C | | Authors: | Qi, P.X, Di Stefano, D.L, Wand, A.J. | | Deposit date: | 1996-07-02 | | Release date: | 1997-07-29 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Solution structure of horse heart ferricytochrome c and detection of redox-related structural changes by high-resolution 1H NMR.
Biochemistry, 35, 1996
|
|
2GZV
 
 | | The cystal structure of the PDZ domain of human PICK1 | | Descriptor: | PRKCA-binding protein | | Authors: | Debreczeni, J.E, Elkins, J.M, Yang, X, Berridge, G, Bray, J, Colebrook, S, Smee, C, Savitsky, P, Gileadi, O, Turnbull, A, von Delft, F, Doyle, D.A, Sundstrom, M, Arrowsmith, C, Weigelt, J, Edwards, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-05-12 | | Release date: | 2006-07-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Structure of PICK1 and other PDZ domains obtained with the help of self-binding C-terminal extensions.
Protein Sci., 16, 2007
|
|
2PZA
 
 | | NAD+ Synthetase from Bacillus anthracis with AMP + PPi and Mg2+ | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | McDonald, H.M, Pruett, P.S, Deivanayagam, C, Protasevich, I.I, Carson, W.M, DeLucas, L.J, Brouillette, W.J, Brouillette, C.G. | | Deposit date: | 2007-05-17 | | Release date: | 2007-07-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural adaptation of an interacting non-native C-terminal helical extension revealed in the crystal structure of NAD(+) synthetase from Bacillus anthracis.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2PNS
 
 | | 1.9 Angstrom resolution crystal structure of a plant cysteine protease Ervatamin-C refinement with cDNA derived amino acid sequence | | Descriptor: | Ervatamin-C, a papain-like plant cysteine protease, PHOSPHATE ION, ... | | Authors: | Ghosh, R, Guha Thakurta, P, Biswas, S, Chakrabarti, C, Dattagupta, J.K. | | Deposit date: | 2007-04-25 | | Release date: | 2007-06-19 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A thermostable cysteine protease precursor from a tropical plant contains an unusual C-terminal propeptide: cDNA cloning, sequence comparison and molecular modeling studies.
Biochem.Biophys.Res.Commun., 362, 2007
|
|
2OT4
 
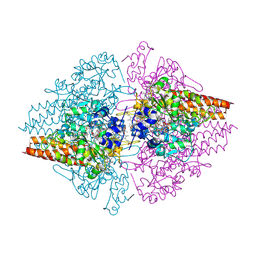 | | Structure of a hexameric multiheme c nitrite reductase from the extremophile bacterium Thiolkalivibrio nitratireducens | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, ... | | Authors: | Polyakov, K.M, Boyko, K.M, Slutsky, A, Tikhonova, T.V, Antipov, A.N, Zvyagilskaya, R.A, Popov, A.N, Lamzin, V.S, Bourenkov, G.P, Popov, V.O. | | Deposit date: | 2007-02-07 | | Release date: | 2008-04-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High-resolution structural analysis of a novel octaheme cytochrome c nitrite reductase from the haloalkaliphilic bacterium Thioalkalivibrio nitratireducens.
J.Mol.Biol., 389, 2009
|
|
1IRW
 
 | |
4KIC
 
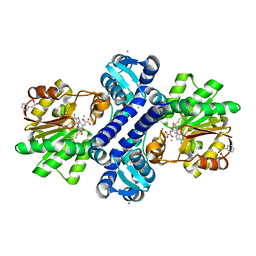 | | Crystal structure of methyltransferase from Streptomyces hygroscopicus complexed with S-adenosyl-L-methionine and phenylpyruvic acid | | Descriptor: | (2R)-2-hydroxy-3-phenylpropanoic acid, 3-PHENYLPYRUVIC ACID, CALCIUM ION, ... | | Authors: | Liu, Y.C, Zou, X.W, Chan, H.C, Huang, C.J, Li, T.L. | | Deposit date: | 2013-05-02 | | Release date: | 2014-05-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structure and mechanism of a nonhaem-iron SAM-dependent C-methyltransferase and its engineering to a hydratase and an O-methyltransferase
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4KIB
 
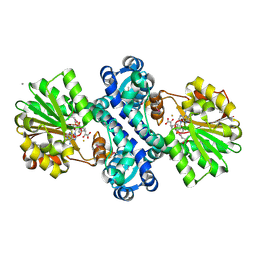 | | Crystal structure of methyltransferase from Streptomyces hygroscopicus complexed with S-adenosyl-L-homocysteine and methylphenylpyruvic acid | | Descriptor: | (2R)-2-hydroxy-3-phenylpropanoic acid, (3R)-2-oxo-3-phenylbutanoic acid, CALCIUM ION, ... | | Authors: | Liu, Y.C, Zou, X.W, Chan, H.C, Huang, C.J, Li, T.L. | | Deposit date: | 2013-05-02 | | Release date: | 2014-05-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and mechanism of a nonhaem-iron SAM-dependent C-methyltransferase and its engineering to a hydratase and an O-methyltransferase
Acta Crystallogr.,Sect.D, 70, 2014
|
|
2N9I
 
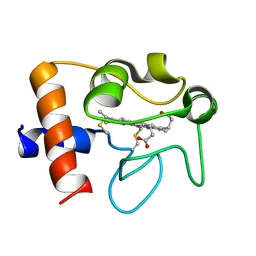 | | Solution structure of reduced human cytochrome c | | Descriptor: | Cytochrome c, HEME C | | Authors: | Imai, M, Saio, T, Kumeta, H, Uchida, T, Inagaki, F, Ishimori, K. | | Deposit date: | 2015-11-24 | | Release date: | 2016-02-17 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Investigation of the redox-dependent modulation of structure and dynamics in human cytochrome c
Biochem.Biophys.Res.Commun., 469, 2016
|
|
