1BWV
 
 | | Activated Ribulose 1,5-Bisphosphate Carboxylase/Oxygenase (RUBISCO) Complexed with the Reaction Intermediate Analogue 2-Carboxyarabinitol 1,5-Bisphosphate | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, PROTEIN (RIBULOSE BISPHOSPHATE CARBOXYLASE) | | Authors: | Sugawara, H, Yamamoto, H, Shibata, N, Inoue, T, Miyake, C, Yokota, A, Kai, Y. | | Deposit date: | 1998-09-29 | | Release date: | 1999-09-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of carboxylase reaction-oriented ribulose 1, 5-bisphosphate carboxylase/oxygenase from a thermophilic red alga, Galdieria partita.
J.Biol.Chem., 274, 1999
|
|
1BWW
 
 | | BENCE-JONES IMMUNOGLOBULIN REI VARIABLE PORTION, T39K MUTANT | | Descriptor: | PROTEIN (IG KAPPA CHAIN V-I REGION REI) | | Authors: | Uson, I, Pohl, E, Schneider, T.R, Dauter, Z, Schmidt, A, Fritz, H.J, Sheldrick, G.M. | | Deposit date: | 1998-09-29 | | Release date: | 1998-10-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 1.7 A structure of the stabilized REIv mutant T39K. Application of local NCS restraints.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1BWX
 
 | | THE SOLUTION STRUCTURE OF HUMAN PARATHYROID HORMONE FRAGMENT 1-39, NMR, 10 STRUCTURES | | Descriptor: | PARATHYROID HORMONE | | Authors: | Marx, U.C, Roesch, P, Adermann, K, Bayer, P, Forssmann, W.-G. | | Deposit date: | 1998-09-29 | | Release date: | 2000-01-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structures of human parathyroid hormone fragments hPTH(1-34) and hPTH(1-39) and bovine parathyroid hormone fragment bPTH(1-37).
Biochem.Biophys.Res.Commun., 267, 2000
|
|
1BWY
 
 | | NMR STUDY OF BOVINE HEART FATTY ACID BINDING PROTEIN | | Descriptor: | PROTEIN (HEART FATTY ACID BINDING PROTEIN) | | Authors: | Lassen, D, Luecke, C, Kveder, M, Mesgarzadeh, A, Schmidt, J.M, Specht, B, Lezius, A, Spener, F, Rueterjans, H. | | Deposit date: | 1998-09-29 | | Release date: | 1998-10-07 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of bovine heart fatty-acid-binding protein with bound palmitic acid, determined by multidimensional NMR spectroscopy.
Eur.J.Biochem., 230, 1995
|
|
1BWZ
 
 | |
1BX0
 
 | | Ferredoxin:nadp+ oxidoreductase (ferredoxin reductase) mutant e312l | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION, PROTEIN (FERREDOXIN:NADP+ OXIDOREDUCTASE), ... | | Authors: | Aliverti, A, Deng, Z, Ravasi, D, Piubelli, L, Karplus, P.A, Zanetti, G. | | Deposit date: | 1998-10-10 | | Release date: | 1998-10-14 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Probing the function of the invariant glutamyl residue 312 in spinach ferredoxin-NADP+ reductase.
J.Biol.Chem., 273, 1998
|
|
1BX1
 
 | | FERREDOXIN:NADP+ OXIDOREDUCTASE (FERREDOXIN REDUCTASE) MUTANT E312Q | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION, PROTEIN (FERREDOXIN:NADP+ OXIDOREDUCTASE), ... | | Authors: | Aliverti, A, Deng, Z, Ravasi, D, Piubelli, L, Karplus, P.A, Zanetti, G. | | Deposit date: | 1998-10-10 | | Release date: | 1998-10-14 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Probing the function of the invariant glutamyl residue 312 in spinach ferredoxin-NADP+ reductase.
J.Biol.Chem., 273, 1998
|
|
1BX2
 
 | | CRYSTAL STRUCTURE OF HLA-DR2 (DRA*0101,DRB1*1501) COMPLEXED WITH A PEPTIDE FROM HUMAN MYELIN BASIC PROTEIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEIN (HLA-DR2) | | Authors: | Smith, K.J, Pyrdol, J, Gauthier, L, Wiley, D.C, Wucherpfennig, K. | | Deposit date: | 1998-10-12 | | Release date: | 1998-10-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of HLA-DR2 (DRA*0101, DRB1*1501) complexed with a peptide from human myelin basic protein.
J.Exp.Med., 188, 1998
|
|
1BX3
 
 | | EFFECTS OF COMMONLY USED CRYOPROTECTANTS ON GLYCOGEN PHOSPHORYLASE ACTIVITY AND STRUCTURE | | Descriptor: | PROTEIN (GLYCOGEN PHOSPHORYLASE B), PYRIDOXAL-5'-PHOSPHATE | | Authors: | Tsitsanou, K.E, Oikonomakos, N.G, Zographos, S.E, Skamnaki, V.T, Gregoriou, M, Watson, K.A, Johnson, L.N, Fleet, G.W.J. | | Deposit date: | 1999-10-13 | | Release date: | 1999-10-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Effects of commonly used cryoprotectants on glycogen phosphorylase activity and structure.
Protein Sci., 8, 1999
|
|
1BX4
 
 | | STRUCTURE OF HUMAN ADENOSINE KINASE AT 1.50 ANGSTROMS | | Descriptor: | ADENOSINE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Mathews, I.I, Erion, M.D, Ealick, S.E. | | Deposit date: | 1998-10-13 | | Release date: | 1999-10-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of human adenosine kinase at 1.5 A resolution.
Biochemistry, 37, 1998
|
|
1BX5
 
 | |
1BX6
 
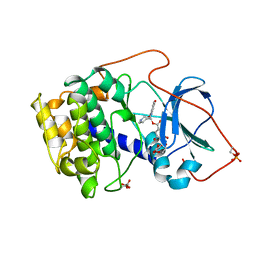 | | CRYSTAL STRUCTURE OF THE POTENT NATURAL PRODUCT INHIBITOR BALANOL IN COMPLEX WITH THE CATALYTIC SUBUNIT OF CAMP-DEPENDENT PROTEIN KINASE | | Descriptor: | BALANOL, CAMP-DEPENDENT PROTEIN KINASE | | Authors: | Narayana, N, Xuong, N.-H, Ten Eyck, L.F, Taylor, S.S. | | Deposit date: | 1998-10-13 | | Release date: | 1999-04-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the potent natural product inhibitor balanol in complex with the catalytic subunit of cAMP-dependent protein kinase.
Biochemistry, 38, 1999
|
|
1BX7
 
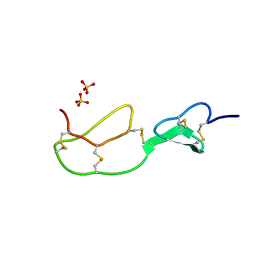 | | HIRUSTASIN FROM HIRUDO MEDICINALIS AT 1.2 ANGSTROMS | | Descriptor: | HIRUSTASIN, SULFATE ION | | Authors: | Uson, I, Sheldrick, G.M, De La Fortelle, E, Bricogne, G, Di Marco, S, Priestle, J.P, Gruetter, M.G, Mittl, P.R.E. | | Deposit date: | 1998-10-14 | | Release date: | 1999-04-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The 1.2 A crystal structure of hirustasin reveals the intrinsic flexibility of a family of highly disulphide-bridged inhibitors.
Structure Fold.Des., 7, 1999
|
|
1BX8
 
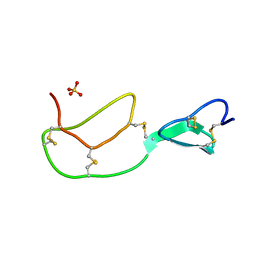 | | HIRUSTASIN FROM HIRUDO MEDICINALIS AT 1.4 ANGSTROMS | | Descriptor: | HIRUSTASIN, SULFATE ION | | Authors: | Uson, I, Sheldrick, G.M, De La Fortelle, E, Bricogne, G, Di Marco, S, Priestle, J.P, Gruetter, M.G, Mittl, P.R.E. | | Deposit date: | 1998-10-14 | | Release date: | 1999-04-27 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The 1.2 A crystal structure of hirustasin reveals the intrinsic flexibility of a family of highly disulphide-bridged inhibitors.
Structure Fold.Des., 7, 1999
|
|
1BX9
 
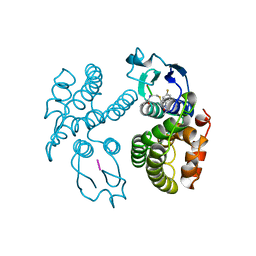 | | GLUTATHIONE S-TRANSFERASE IN COMPLEX WITH HERBICIDE | | Descriptor: | FOE-4053-glutathione conjugate GGL-FOE-GLY, GLUTATHIONE S-TRANSFERASE | | Authors: | Prade, L, Huber, R, Bieseler, B. | | Deposit date: | 1998-10-14 | | Release date: | 1998-10-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of herbicides in complex with their detoxifying enzyme glutathione S-transferase - explanations for the selectivity of the enzyme in plants.
Structure, 6, 1998
|
|
1BXA
 
 | | AMICYANIN REDUCED, PH 4.4, 1.3 ANGSTROMS | | Descriptor: | COPPER (I) ION, PROTEIN (AMICYANIN) | | Authors: | Cunane, L.M, Chen, Z.W, Durley, R.C.E, Mathews, F.S. | | Deposit date: | 1998-10-01 | | Release date: | 1998-10-07 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Molecular basis for interprotein complex-dependent effects on the redox properties of amicyanin.
Biochemistry, 37, 1998
|
|
1BXB
 
 | | XYLOSE ISOMERASE FROM THERMUS THERMOPHILUS | | Descriptor: | XYLOSE ISOMERASE | | Authors: | Chang, C, Park, B.C, Lee, D.-S, Suh, S.W. | | Deposit date: | 1998-10-02 | | Release date: | 1999-02-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of thermostable xylose isomerases from Thermus caldophilus and Thermus thermophilus: possible structural determinants of thermostability.
J.Mol.Biol., 288, 1999
|
|
1BXC
 
 | | XYLOSE ISOMERASE FROM THERMUS CALDOPHILUS | | Descriptor: | XYLOSE ISOMERASE | | Authors: | Chang, C, Park, B.C, Lee, D.-S, Suh, S.W. | | Deposit date: | 1998-10-02 | | Release date: | 1999-02-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of thermostable xylose isomerases from Thermus caldophilus and Thermus thermophilus: possible structural determinants of thermostability.
J.Mol.Biol., 288, 1999
|
|
1BXD
 
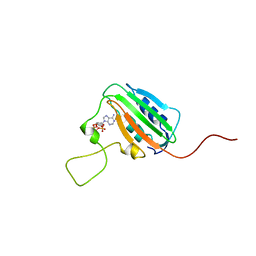 | | NMR STRUCTURE OF THE HISTIDINE KINASE DOMAIN OF THE E. COLI OSMOSENSOR ENVZ | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, PROTEIN (OSMOLARITY SENSOR PROTEIN (ENVZ)) | | Authors: | Tanaka, T, Saha, S.K, Tomomori, C, Ishima, R, Liu, D, Tong, K.I, Park, H, Dutta, R, Qin, L, Swindells, M.B, Yamazaki, T, Ono, A.M, Kainosho, M, Inouye, M, Ikura, M. | | Deposit date: | 1998-10-02 | | Release date: | 1999-10-02 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the histidine kinase domain of the E. coli osmosensor EnvZ.
Nature, 396, 1998
|
|
1BXE
 
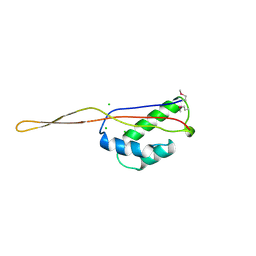 | | RIBOSOMAL PROTEIN L22 FROM THERMUS THERMOPHILUS | | Descriptor: | CHLORIDE ION, PROTEIN (RIBOSOMAL PROTEIN L22) | | Authors: | Unge, J, Aberg, A, Al-Karadaghi, S, Nikulin, A, Nikonov, S, Davydova, N, Nevskaya, N, Garber, M, Liljas, A. | | Deposit date: | 1998-10-02 | | Release date: | 1998-10-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of ribosomal protein L22 from Thermus thermophilus: insights into the mechanism of erythromycin resistance.
Structure, 6, 1998
|
|
1BXG
 
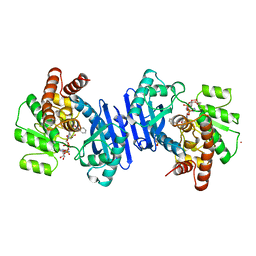 | | PHENYLALANINE DEHYDROGENASE STRUCTURE IN TERNARY COMPLEX WITH NAD+ AND BETA-PHENYLPROPIONATE | | Descriptor: | HYDROCINNAMIC ACID, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PHENYLALANINE DEHYDROGENASE, ... | | Authors: | Vanhooke, J.L, Thoden, J.B, Brunhuber, N.M.W, Blanchard, J.L, Holden, H.M. | | Deposit date: | 1998-10-02 | | Release date: | 1999-05-18 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Phenylalanine dehydrogenase from Rhodococcus sp. M4: high-resolution X-ray analyses of inhibitory ternary complexes reveal key features in the oxidative deamination mechanism.
Biochemistry, 38, 1999
|
|
1BXH
 
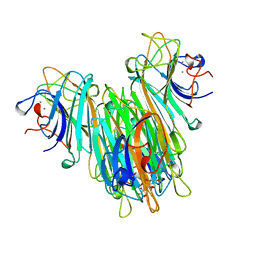 | | CONCANAVALIN A COMPLEXED TO METHYL ALPHA1-2 MANNOBIOSIDE | | Descriptor: | CALCIUM ION, Concanavalin-A, MANGANESE (II) ION, ... | | Authors: | Moothoo, D.N, Canaan, B, Field, R.A, Naismith, J.H. | | Deposit date: | 1998-10-02 | | Release date: | 1998-10-07 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Man alpha1-2 Man alpha-OMe-concanavalin A complex reveals a balance of forces involved in carbohydrate recognition.
Glycobiology, 9, 1999
|
|
1BXI
 
 | | CRYSTAL STRUCTURE OF THE ESCHERICHIA COLI COLICIN E9 DNASE DOMAIN WITH ITS COGNATE IMMUNITY PROTEIN IM9 | | Descriptor: | NICKEL (II) ION, PHOSPHATE ION, PROTEIN (COLICIN E9 IMMUNITY PROTEIN), ... | | Authors: | Kuhlmann, U.C, Kleanthous, C, James, R, Moore, G.R, Hemmings, A.M. | | Deposit date: | 1998-10-04 | | Release date: | 1999-10-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structure of the E.Coli Colicin E9 DNase Domain With its Cognate Immunity Protein Im9
Thesis, 1998
|
|
1BXK
 
 | | DTDP-GLUCOSE 4,6-DEHYDRATASE FROM E. COLI | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PROTEIN (DTDP-GLUCOSE 4,6-DEHYDRATASE) | | Authors: | Thoden, J.B, Hegeman, A.D, Frey, P.A, Holden, H.M. | | Deposit date: | 1998-10-05 | | Release date: | 1998-10-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular Structure of Dtdp-Glucose 4,6-Dehydratase from E. Coli
Protein Sci.
|
|
1BXL
 
 | | STRUCTURE OF BCL-XL/BAK PEPTIDE COMPLEX, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | BAK PEPTIDE, BCL-XL | | Authors: | Sattler, M, Liang, H, Nettesheim, D, Meadows, R.P, Harlan, J.E, Eberstadt, M, Yoon, H, Shuker, S.B, Chang, B.S, Minn, A.J, Thompson, C.B, Fesik, S.W. | | Deposit date: | 1996-10-16 | | Release date: | 1997-10-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of Bcl-xL-Bak peptide complex: recognition between regulators of apoptosis.
Science, 275, 1997
|
|
