6P6Q
 
 | | HCV NS3/4A protease domain of genotype 1a3a chimera in complex with grazoprevir | | Descriptor: | (1aR,5S,8S,10R,22aR)-5-tert-butyl-N-{(1R,2S)-1-[(cyclopropylsulfonyl)carbamoyl]-2-ethenylcyclopropyl}-14-methoxy-3,6-di oxo-1,1a,3,4,5,6,9,10,18,19,20,21,22,22a-tetradecahydro-8H-7,10-methanocyclopropa[18,19][1,10,3,6]dioxadiazacyclononadec ino[11,12-b]quinoxaline-8-carboxamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Non-structural protein 4A,Serine protease NS3, ... | | Authors: | Timm, J, Schiffer, C.A. | | Deposit date: | 2019-06-04 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Molecular mechanism of pan-genotypic HCV NS3/4A protease inhibition by glecaprevir and characterization of genotype-specific structural differences
To Be Published
|
|
8DK6
 
 | | Structure of hepatitis C virus envelope N-terminal truncated glycoprotein 2 (E2) (residues 456-713) from J6 genotype | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2A12 Fab Heavy chain, 2A12 Fab light chain, ... | | Authors: | Kumar, A, Rohe, T, Elrod, E.J, Khan, A.G, Dearborn, A.D, Kissinger, R, Grakoui, A, Marcotrigiano, J. | | Deposit date: | 2022-07-03 | | Release date: | 2023-03-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Regions of hepatitis C virus E2 required for membrane association.
Nat Commun, 14, 2023
|
|
6UL9
 
 | | Crystal structure of human GAC in complex with inhibitor UPGL00023 | | Descriptor: | 2-phenyl-N-{5-[(1-{5-[(phenylacetyl)amino]-1,3,4-thiadiazol-2-yl}azetidin-3-yl)oxy]-1,3,4-thiadiazol-2-yl}acetamide, Glutaminase kidney isoform, mitochondrial | | Authors: | Huang, Q.Q, Cerione, R.A. | | Deposit date: | 2019-10-07 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of human GAC in complex with inhibitor UPGL00012
To Be Published
|
|
8DZ8
 
 | |
6M15
 
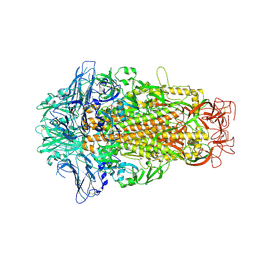 | | Cryo-EM structures of HKU2 spike glycoproteins | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Wang, X, Yu, J, Qiao, S, Guo, R. | | Deposit date: | 2020-02-24 | | Release date: | 2020-05-27 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (2.38 Å) | | Cite: | Cryo-EM structures of HKU2 and SADS-CoV spike glycoproteins provide insights into coronavirus evolution.
Nat Commun, 11, 2020
|
|
6UNJ
 
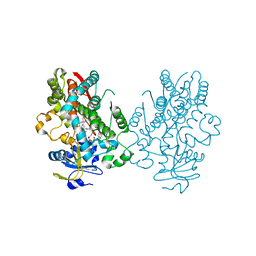 | | Human CYP3A4 bound to an inhibitor | | Descriptor: | Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE, tert-butyl [(2R)-1-(naphthalen-1-yl)-3-{[(2S)-1-oxo-3-phenyl-1-{[2-(pyridin-3-yl)ethyl]amino}propan-2-yl]sulfanyl}propan-2-yl]carbamate | | Authors: | Sevrioukova, I. | | Deposit date: | 2019-10-12 | | Release date: | 2020-02-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | An increase in side-group hydrophobicity largely improves the potency of ritonavir-like inhibitors of CYP3A4.
Bioorg.Med.Chem., 28, 2020
|
|
5PXZ
 
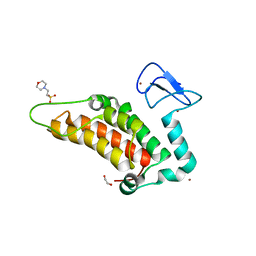 | | PanDDA analysis group deposition -- Crystal Structure of SP100 after initial refinement with no ligand modelled (structure 59) | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Nuclear autoantigen Sp-100, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-08 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
7ML8
 
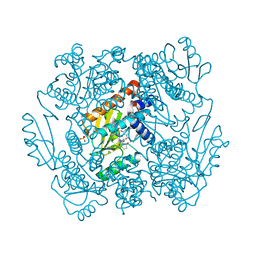 | | The crystal structure of I38T mutant PA endonuclease (2009/H1N1/CALIFORNIA) in complex with SJ001023038 | | Descriptor: | 5-hydroxy-N-[2-(4-hydroxy-3-methoxyphenyl)ethyl]-6-oxo-2-phenyl-1,6-dihydropyrimidine-4-carboxamide, GLUTAMIC ACID, Hexa Vinylpyrrolidone K15, ... | | Authors: | Cuypers, M.G, Slavish, P.J, Yun, M.K, Dubois, R, Rankovic, Z, White, S.W. | | Deposit date: | 2021-04-27 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of I38T mutant PA endonuclease (2009/H1N1/CALIFORNIA) in complex with St Jude compound AC067-19
To Be Published
|
|
6P6Z
 
 | | HCV NS3/4A protease domain of genotype 4a with an extended linker in complex with glecaprevir | | Descriptor: | (3aR,7S,10S,12R,21E,24aR)-7-tert-butyl-N-[(1R,2R)-2-(difluoromethyl)-1-{[(1-methylcyclopropyl)sulfonyl]carbamoyl}cyclop ropyl]-20,20-difluoro-5,8-dioxo-2,3,3a,5,6,7,8,11,12,20,23,24a-dodecahydro-1H,10H-9,12-methanocyclopenta[18,19][1,10,17, 3,6]trioxadiazacyclononadecino[11,12-b]quinoxaline-10-carboxamide, Non-structural protein 4A,Serine protease NS3, ... | | Authors: | Timm, J, Schiffer, C.A. | | Deposit date: | 2019-06-04 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.294 Å) | | Cite: | Molecular mechanism of pan-genotypic HCV NS3/4A protease inhibition by glecaprevir and characterization of genotype-specific structural differences
To Be Published
|
|
5PYG
 
 | | PanDDA analysis group deposition -- Crystal Structure of SP100 after initial refinement with no ligand modelled (structure 76) | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Nuclear autoantigen Sp-100, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-08 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
8E15
 
 | | A computationally stabilized hMPV F protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, F1 protein with Fibritin peptide, F2 protein, ... | | Authors: | Huang, J, Gonzalez, K, Mousa, J, Strauch, E. | | Deposit date: | 2022-08-09 | | Release date: | 2023-04-12 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | A general computational design strategy for stabilizing viral class I fusion proteins.
Nat Commun, 15, 2024
|
|
6ZUO
 
 | | Human RIO1(kd)-StHA late pre-40S particle, structural state A (pre 18S rRNA cleavage) | | Descriptor: | 40S ribosomal protein S10, 40S ribosomal protein S11, 40S ribosomal protein S12, ... | | Authors: | Plassart, L, Shayan, R, Plisson-Chastang, C. | | Deposit date: | 2020-07-23 | | Release date: | 2021-05-12 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The final step of 40S ribosomal subunit maturation is controlled by a dual key lock.
Elife, 10, 2021
|
|
6M2F
 
 | |
6UD1
 
 | | Spectroscopic and structural characterization of a genetically encoded direct sensor for protein-ligand interactions | | Descriptor: | CHLORIDE ION, Streptavidin | | Authors: | Mills, J.H, Gleason, P.R, Simmons, C.R, Henderson, J.N, Kartchner, B.K. | | Deposit date: | 2019-09-18 | | Release date: | 2021-02-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Origins of Altered Spectroscopic Properties upon Ligand Binding in Proteins Containing a Fluorescent Noncanonical Amino Acid.
Biochemistry, 60, 2021
|
|
6P74
 
 | | OLD nuclease from Thermus Scotoductus | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, PLATINUM (II) ION, Putative ATP-dependent endonuclease of the OLD family, ... | | Authors: | Chappie, J.S, Schiltz, C.J. | | Deposit date: | 2019-06-04 | | Release date: | 2020-01-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The full-length structure of Thermus scotoductus OLD defines the ATP hydrolysis properties and catalytic mechanism of Class 1 OLD family nucleases.
Nucleic Acids Res., 48, 2020
|
|
8EBU
 
 | | XPC release from Core7-XPA-DNA (Cy5) | | Descriptor: | DNA repair protein complementing XP-A cells, DNA repair protein complementing XP-C cells, DNA1, ... | | Authors: | Kim, J, Yang, W. | | Deposit date: | 2022-08-31 | | Release date: | 2023-04-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Lesion recognition by XPC, TFIIH and XPA in DNA excision repair.
Nature, 617, 2023
|
|
6M33
 
 | |
5PUW
 
 | | PanDDA analysis group deposition -- Crystal Structure of BRD1 after initial refinement with no ligand modelled (structure 210) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 1, SODIUM ION | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
7A9J
 
 | |
6PEH
 
 | | Crystal structure of rabbit monoclonal anti-HIV antibody 1C2 | | Descriptor: | 1C2 Fab Heavy Chain, 1C2 Fab Light Chain, SULFATE ION | | Authors: | Liban, T, Pancera, M. | | Deposit date: | 2019-06-20 | | Release date: | 2019-11-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.296 Å) | | Cite: | Vaccination with Glycan-Modified HIV NFL Envelope Trimer-Liposomes Elicits Broadly Neutralizing Antibodies to Multiple Sites of Vulnerability.
Immunity, 51, 2019
|
|
6U1L
 
 | | Structure of two-domain translational regulator Yih1 reveals a possible mechanism of action | | Descriptor: | Protein IMPACT homolog | | Authors: | Harjes, E, Jameson, G.B, Edwards, P.J.B, Goroncy, A.K, Loo, T, Norris, G.E. | | Deposit date: | 2019-08-16 | | Release date: | 2021-02-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Experimentally based structural model of Yih1 provides insight into its function in controlling the key translational regulator Gcn2.
Febs Lett., 595, 2021
|
|
7MN5
 
 | | Structure of the HER2/HER3/NRG1b Heterodimer Extracellular Domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Isoform 6 of Pro-neuregulin-1, ... | | Authors: | Diwanji, D, Trenker, R, Verba, K.A, Jura, N. | | Deposit date: | 2021-04-30 | | Release date: | 2021-10-27 | | Last modified: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structures of the HER2-HER3-NRG1 beta complex reveal a dynamic dimer interface.
Nature, 600, 2021
|
|
8E7C
 
 | |
6PEW
 
 | |
6M3C
 
 | | hAPC-h1573 Fab complex | | Descriptor: | Vitamin K-dependent protein C heavy chain, Vitamin K-dependent protein C light chain, h1573 Fab H chain, ... | | Authors: | Wang, X, Wang, D, Zhao, X, Egner, U. | | Deposit date: | 2020-03-03 | | Release date: | 2020-07-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Targeted inhibition of activated protein C by a non-active-site inhibitory antibody to treat hemophilia.
Nat Commun, 11, 2020
|
|
