8V04
 
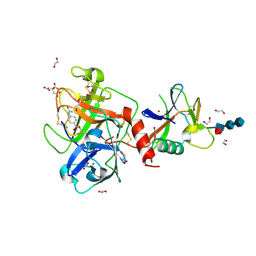 | | High resolution TMPRSS2 structure following acylation by nafamostat | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 4-carbamimidamidobenzoic acid, ... | | Authors: | Fraser, B.J, Dong, A, Kutera, M, Seitova, A, Li, Y, Hutchinson, A, Edwards, A, Benard, F, Levon, H, Arrowsmith, C. | | Deposit date: | 2023-11-16 | | Release date: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | High resolution TMPRSS2 structure following acylation by nafamostat
To Be Published
|
|
8V02
 
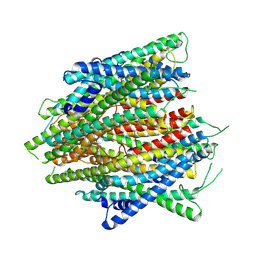 | | AaegOR10 structure bound to o-cresol | | Descriptor: | Odorant receptor OR10, Odorant receptor Orco, o-cresol | | Authors: | Zhao, J, del Marmol, J. | | Deposit date: | 2023-11-16 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of odor sensing by insect heteromeric odorant receptors.
Science, 384, 2024
|
|
8V01
 
 | |
8V00
 
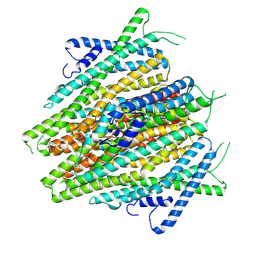 | | AaegOR10 apo structure | | Descriptor: | Odorant receptor OR10, Odorant receptor Orco | | Authors: | Zhao, J, del Marmol, J. | | Deposit date: | 2023-11-16 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Structural basis of odor sensing by insect heteromeric odorant receptors.
Science, 384, 2024
|
|
8UZY
 
 | |
8UZX
 
 | |
8UZW
 
 | |
8UZU
 
 | | Crystal structure of Shewanella benthica Group 1 truncated hemoglobin L80A C51S C71S variant | | Descriptor: | CYANIDE ION, Group 1 truncated hemoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Schultz, T.D, Martinez, J.E, Siegler, M.A, Schlessman, J.L, Lecomte, J.T.J. | | Deposit date: | 2023-11-16 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Shewanella benthica Group 1 truncated hemoglobin L80A C51S C71S variant
To Be Published
|
|
8UZO
 
 | |
8UZN
 
 | |
8UZM
 
 | |
8UZL
 
 | | Designed Transmembrane beta-barrel- TMB10_163 | | Descriptor: | Designed Transmembrane beta-barrel TMB10_163, HEXANE-1,6-DIOL | | Authors: | Bera, A.K, Lemma, S.B, Kang, A, Baker, D. | | Deposit date: | 2023-11-15 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Sculpting conducting nanopore size and shape through de novo protein design
Science, 2024
|
|
8UZK
 
 | |
8UZJ
 
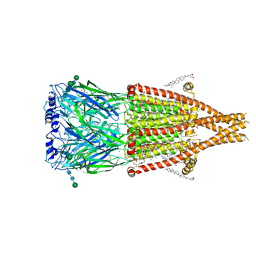 | | Alpha7-nicotinic acetylcholine receptor bound to epibatidine and ivermectin | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, (2aE,4E,5'S,6S,6'R,7S,8E,11R,13R,15S,17aR,20R,20aR,20bS)-6'-[(2S)-butan-2-yl]-20,20b-dihydroxy-5',6,8,19-tetramethyl-17 -oxo-3',4',5',6,6',10,11,14,15,17,17a,20,20a,20b-tetradecahydro-2H,7H-spiro[11,15-methanofuro[4,3,2-pq][2,6]benzodioxacy clooctadecine-13,2'-pyran]-7-yl 2,6-dideoxy-4-O-(2,6-dideoxy-3-O-methyl-alpha-L-arabino-hexopyranosyl)-3-O-methyl-alpha-L-arabino-hexopyranoside, (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, ... | | Authors: | Burke, S.M, Noviello, C.M, Hibbs, R.E. | | Deposit date: | 2023-11-15 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Structural mechanisms of alpha 7 nicotinic receptor allosteric modulation and activation.
Cell, 187, 2024
|
|
8UZI
 
 | |
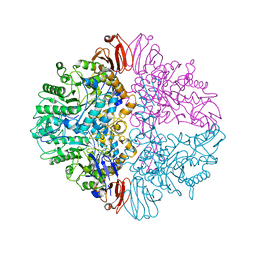
8UZH
 
 | |
8UZD
 
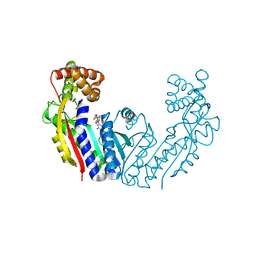 | |
8UZB
 
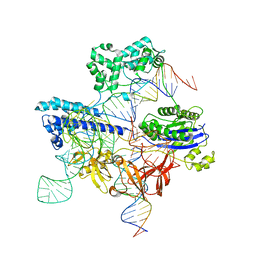 | | Cryo-EM structure of iGeoCas9 in complex with sgRNA and target DNA | | Descriptor: | CRISPR-associated endonuclease Cas9, Non-target strand DNA, RNA (107-MER), ... | | Authors: | Eggers, A.R, Soczek, K.M, Tuck, O.T, Doudna, J.A. | | Deposit date: | 2023-11-14 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.63 Å) | | Cite: | Rapid DNA unwinding accelerates genome editing by engineered CRISPR-Cas9.
Cell, 187, 2024
|
|
8UZA
 
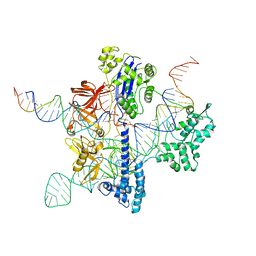 | | Cryo-EM structure of GeoCas9 in complex with sgRNA and target DNA | | Descriptor: | CRISPR-associated endonuclease Cas9, Non-target strand DNA, Target strand DNA, ... | | Authors: | Eggers, A.R, Soczek, K.M, Tuck, O.T, Doudna, J.A. | | Deposit date: | 2023-11-14 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Rapid DNA unwinding accelerates genome editing by engineered CRISPR-Cas9.
Cell, 187, 2024
|
|
8UZ8
 
 | |
8UZ7
 
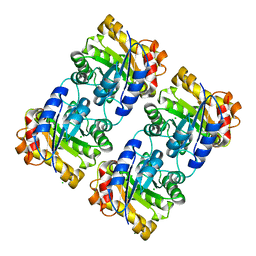 | | Crystal structure of a novel triose phosphate isomerase identified on the shrimp transcriptome | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Triosephosphate isomerase | | Authors: | Sotelo-Mundo, R.R, Gomez-Yanes, A.C, Lopez-Zavala, A.A, Lopez-Garcia, J.A, Ochoa-Leyva, A. | | Deposit date: | 2023-11-14 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a novel triose phosphate isomerase from shrimp
To Be Published
|
|
8UZ6
 
 | |
8UZ5
 
 | |
8UZ4
 
 | |
8UZ1
 
 | | Straight actin filament from Arp2/3 branch junction sample (ADP-BeFx) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Chavali, S.S, Chou, S.Z, Sindelar, C.V. | | Deposit date: | 2023-11-14 | | Release date: | 2024-01-31 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures reveal how phosphate release from Arp3 weakens actin filament branches formed by Arp2/3 complex.
Nat Commun, 15, 2024
|
|
