1QQB
 
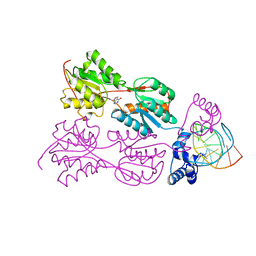 | | PURINE REPRESSOR MUTANT-HYPOXANTHINE-PALINDROMIC OPERATOR COMPLEX | | Descriptor: | 5'-D(*TP*AP*CP*GP*CP*AP*AP*TP*CP*GP*AP*TP*TP*GP*CP*GP*T)-3', HYPOXANTHINE, PURINE NUCLEOTIDE SYNTHESIS REPRESSOR | | Authors: | Glasfeld, A, Koehler, A.N, Schumacher, M.A, Brennan, R.G. | | Deposit date: | 1999-06-01 | | Release date: | 1999-06-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The role of lysine 55 in determining the specificity of the purine repressor for its operators through minor groove interactions.
J.Mol.Biol., 291, 1999
|
|
3A5I
 
 | |
1QSC
 
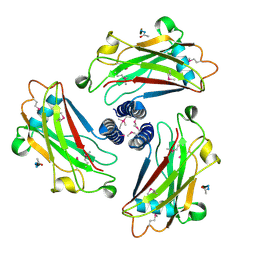 | | CRYSTAL STRUCTURE OF THE TRAF DOMAIN OF TRAF2 IN A COMPLEX WITH A PEPTIDE FROM THE CD40 RECEPTOR | | Descriptor: | CD40 RECEPTOR, TNF RECEPTOR ASSOCIATED FACTOR 2 | | Authors: | McWhirter, S.M, Pullen, S.S, Holton, J.M, Crute, J.J, Kehry, M.R, Alber, T. | | Deposit date: | 1999-06-20 | | Release date: | 1999-08-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystallographic analysis of CD40 recognition and signaling by human TRAF2.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
2DT4
 
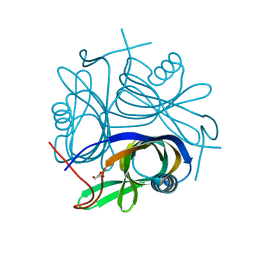 | | Crystal structure of Pyrococcus horikoshii a plant- and prokaryote-conserved (PPC) protein at 1.60 resolution | | Descriptor: | GLYCEROL, Hypothetical protein PH0802 | | Authors: | Lin, L, Nakano, H, Uchiyama, S, Fujimoto, S, Matsunaga, S, Nakamura, S. | | Deposit date: | 2006-07-10 | | Release date: | 2007-05-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of Pyrococcus horikoshii PPC protein at 1.60 A resolution
Proteins, 67, 2007
|
|
3FK0
 
 | | E. coli EPSP synthase (TIPS mutation) liganded with S3P | | Descriptor: | 3-phosphoshikimate 1-carboxyvinyltransferase, FORMIC ACID, SHIKIMATE-3-PHOSPHATE | | Authors: | Schonbrunn, E. | | Deposit date: | 2008-12-15 | | Release date: | 2009-02-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis of Glyphosate Resistance Resulting from the Double Mutation Thr97 -> Ile and Pro101 -> Ser in 5-Enolpyruvylshikimate-3-phosphate Synthase from Escherichia coli.
J.Biol.Chem., 284, 2009
|
|
387D
 
 | | RNA Pseudoknot with 3D Domain Swapping | | Descriptor: | RNA Pseudoknot | | Authors: | Lietzke, S.E, Kundrot, C.E, Barnes, C.L. | | Deposit date: | 1998-04-14 | | Release date: | 2003-08-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The Structure of an RNA Pseudoknot Shows 3D Domain Swapping
Structure, Motion, Interaction and Expression of Biological Macromolecules, The Proceedings of the Tenth Conversation held at The University-SUNY, Albany NY, June 17-21, 1997, 10, 1998
|
|
1QQ6
 
 | | STRUCTURE OF L-2-HALOACID DEHALOGENASE FROM XANTHOBACTER AUTOTROPHICUS WITH CHLOROACETIC ACID COVALENTLY BOUND | | Descriptor: | CHLORIDE ION, PROTEIN (L-2-HALOACID DEHALOGENASE) | | Authors: | Ridder, I.S, Rozeboom, H.J, Kalk, K.H, Dijkstra, B.W. | | Deposit date: | 1999-06-11 | | Release date: | 1999-10-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of intermediates in the dehalogenation of haloalkanoates by L-2-haloacid dehalogenase.
J.Biol.Chem., 274, 1999
|
|
3NT0
 
 | |
1QGD
 
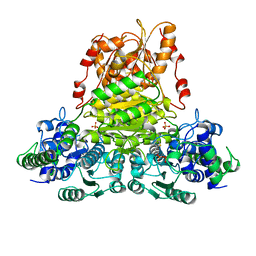 | |
3FPX
 
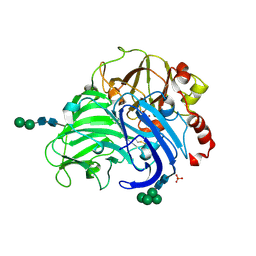 | | Native fungus laccase from Trametes hirsuta | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, ... | | Authors: | Polyakov, K.M, Fedorova, T.V, Stepanova, E.V, Cherkashin, E.A, Kurzeev, S.A, Strokopytov, B.V, Lamzin, V.S, Koroleva, O.V. | | Deposit date: | 2009-01-06 | | Release date: | 2009-01-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of native laccase from Coriolus hirsutus at 1.8 A resolution
To be Published
|
|
1LDY
 
 | |
1Q96
 
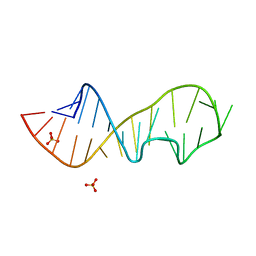 | | Crystal structure of a mutant of the sarcin/ricin domain from rat 28S rRNA | | Descriptor: | SULFATE ION, sarcin/ricin 28S rRNA | | Authors: | Correll, C.C, Beneken, J, Plantinga, M.J, Lubbers, M, Chan, Y.L. | | Deposit date: | 2003-08-22 | | Release date: | 2003-11-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The common and distinctive features of the bulged-G motif based on a 1.04 A resolution RNA structure
Nucleic Acids Res., 31, 2003
|
|
3F5X
 
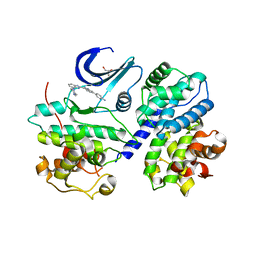 | | CDK-2-Cyclin complex with indazole inhibitor 9 bound at its active site | | Descriptor: | 4-{3-[7-(4-methylpiperazin-1-yl)-1H-benzimidazol-2-yl]-1H-indazol-6-yl}aniline, Cell division protein kinase 2, Cyclin-A2, ... | | Authors: | Kiefer, J.R, Day, J.E, Caspers, N.L, Mathis, K.J, Kretzmer, K.K, Weinberg, R.A, Reitz, B.A, Stegeman, R.A, Trujillo, J.I, Huang, W, Thorarensen, A, Xing, L, Wrightstone, A, Christine, L, Compton, R, Li, X. | | Deposit date: | 2008-11-04 | | Release date: | 2009-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 2-(6-Phenyl-1H-indazol-3-yl)-1H-benzo[d]imidazoles: Design and synthesis of a potent and isoform selective PKC-zeta inhibitor.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
1Q8L
 
 | | Second Metal Binding Domain of the Menkes ATPase | | Descriptor: | Copper-transporting ATPase 1 | | Authors: | Jones, C.E, Daly, N.L, Cobine, P.A, Craik, D.J, Dameron, C.T. | | Deposit date: | 2003-08-21 | | Release date: | 2004-01-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and metal binding studies of the second copper binding domain of the Menkes ATPase.
J.Struct.Biol., 143, 2003
|
|
3AC0
 
 | | Crystal structure of Beta-glucosidase from Kluyveromyces marxianus in complex with glucose | | Descriptor: | Beta-glucosidase I, beta-D-glucopyranose | | Authors: | Yoshida, E, Hidaka, M, Fushinobu, S, Katayama, T, Kumagai, H. | | Deposit date: | 2009-12-25 | | Release date: | 2010-08-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Role of a PA14 domain in determining substrate specificity of a glycoside hydrolase family 3 beta-glucosidase from Kluyveromyces marxianus.
Biochem.J., 431, 2010
|
|
3FC6
 
 | | hRXRalpha & mLXRalpha with an indole Pharmacophore, SB786875 | | Descriptor: | Nr1h3 protein, RETINOIC ACID, Retinoic acid receptor RXR-alpha, ... | | Authors: | Washburn, D.G, Hoang, T.H, Campobasso, N, Smallwood, A, Parks, D.J, Webb, C.L, Frank, K, Nord, M, Duraiswami, C, Evans, C, Jaye, M, Thompson, S.K. | | Deposit date: | 2008-11-21 | | Release date: | 2009-02-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.063 Å) | | Cite: | Synthesis and SAR of potent LXR agonists containing an indole pharmacophore.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
1QK9
 
 | | The solution structure of the domain from MeCP2 that binds to methylated DNA | | Descriptor: | METHYL-CPG-BINDING PROTEIN 2 | | Authors: | Wakefield, R.I.D, Smith, B.O, Nan, X, Free, A, Soteriou, A, Uhrin, D, Bird, A.P, Barlow, P.N. | | Deposit date: | 1999-07-12 | | Release date: | 1999-10-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Solution Structure of the Domain from Mecp2 that Binds to Methylated DNA
J.Mol.Biol., 291, 1999
|
|
2WKW
 
 | | Alcaligenes esterase complexed with product analogue | | Descriptor: | CARBOXYLESTERASE, GLYCEROL, SULFATE ION, ... | | Authors: | Bourne, P.C, Isupov, M.N, Littlechild, J.A. | | Deposit date: | 2009-06-18 | | Release date: | 2009-06-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | The Atomic-Resolution Structure of a Novel Bacterial Esterase.
Structure, 8, 2000
|
|
3F9N
 
 | | Crystal structure of chk1 kinase in complex with inhibitor 38 | | Descriptor: | 3-(3-chlorophenyl)-2-({(1S)-1-[(6S)-2,8-diazaspiro[5.5]undec-2-ylcarbonyl]pentyl}sulfanyl)quinazolin-4(3H)-one, SULFATE ION, Serine/threonine-protein kinase Chk1 | | Authors: | Yan, Y, Munshi, S, Ikuta, M. | | Deposit date: | 2008-11-14 | | Release date: | 2009-01-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Development of thioquinazolinones, allosteric Chk1 kinase inhibitors.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
1Q9D
 
 | | Fructose-1,6-bisphosphatase Complexed with a New Allosteric Site Inhibitor (I-State) | | Descriptor: | 3-(4-HYDROXYBENZYL)-2-[1-({[2-(4-HYDROXYPHENYL)ETHYL]AMINO}CARBONYL)BUTYL]-4-OXO-3,6,11,11A-TETRAHYDRO-4H-PYRAZINO[1,2-B]ISOQUINOLIN-2-IUM-1-OLATE, 6-O-phosphono-beta-D-fructofuranose, Fructose-1,6-bisphosphatase, ... | | Authors: | Honzatko, R.B, Choe, J.Y. | | Deposit date: | 2003-08-25 | | Release date: | 2003-12-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Inhibition of fructose-1,6-bisphosphatase by a new class of allosteric effectors
J.Biol.Chem., 278, 2003
|
|
1QQT
 
 | | METHIONYL-TRNA SYNTHETASE FROM ESCHERICHIA COLI | | Descriptor: | METHIONYL-TRNA SYNTHETASE, ZINC ION | | Authors: | Mechulam, Y, Schmitt, E, Maveyraud, L, Zelwer, C, Nureki, O, Yokoyama, S, Konno, M, Blanquet, S. | | Deposit date: | 1999-06-08 | | Release date: | 2000-01-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal structure of Escherichia coli methionyl-tRNA synthetase highlights species-specific features.
J.Mol.Biol., 294, 1999
|
|
2UX3
 
 | | X-ray high resolution structure of the photosynthetic reaction center from Rb. sphaeroides at pH 9 in the neutral state | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, CARDIOLIPIN, ... | | Authors: | Koepke, J, Diehm, R, Fritzsch, G. | | Deposit date: | 2007-03-26 | | Release date: | 2007-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Ph Modulates the Quinone Position in the Photosynthetic Reaction Center from Rhodobacter Sphaeroides in the Neutral and Charge Separated States.
J.Mol.Biol., 371, 2007
|
|
2UWU
 
 | | X-ray high resolution structure of the photosynthetic reaction center from Rb. sphaeroides at pH 6.5 in the neutral state, 2nd dataset | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, CARDIOLIPIN, ... | | Authors: | Koepke, J, Diehm, R, Fritzsch, G. | | Deposit date: | 2007-03-23 | | Release date: | 2007-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Ph Modulates the Quinone Position in the Photosynthetic Reaction Center from Rhodobacter Sphaeroides in the Neutral and Charge Separated States.
J.Mol.Biol., 371, 2007
|
|
3O6W
 
 | | Crystal structure of monomeric KlHxk1 in crystal form VIII (open state) | | Descriptor: | GLYCEROL, Hexokinase, PHOSPHATE ION | | Authors: | Kuettner, E.B, Kettner, K, Keim, A, Kriegel, T.M, Strater, N. | | Deposit date: | 2010-07-29 | | Release date: | 2010-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal Structure of Hexokinase KlHxk1 of Kluyveromyces lactis: A MOLECULAR BASIS FOR UNDERSTANDING THE CONTROL OF YEAST HEXOKINASE FUNCTIONS VIA COVALENT MODIFICATION AND OLIGOMERIZATION.
J.Biol.Chem., 285, 2010
|
|
1UE4
 
 | | Crystal structure of d(GCGAAAGC) | | Descriptor: | 5'-D(*GP*CP*GP*AP*AP*AP*GP*C)-3', MAGNESIUM ION | | Authors: | Sunami, T, Kondo, J, Hirao, I, Watanabe, K, Miura, K, Takenaka, A. | | Deposit date: | 2003-05-09 | | Release date: | 2004-03-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structures of d(GCGAAGC) and d(GCGAAAGC) (tetragonal form): a switching of partners of the sheared G.A pairs to form a functional G.AxA.G crossing.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
