6NUQ
 
 | | Stat3 Core in complex with compound SI109 | | Descriptor: | Signal transducer and activator of transcription 3, [(2-{[(5S,8S,10aR)-3-acetyl-8-({(2S)-5-amino-1-[(diphenylmethyl)amino]-1,5-dioxopentan-2-yl}carbamoyl)-6-oxodecahydropyrrolo[1,2-a][1,5]diazocin-5-yl]carbamoyl}-1H-indol-5-yl)(difluoro)methyl]phosphonic acid (non-preferred name) | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2019-02-01 | | Release date: | 2019-12-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | A Potent and Selective Small-Molecule Degrader of STAT3 Achieves Complete Tumor Regression In Vivo.
Cancer Cell, 36, 2019
|
|
2RIM
 
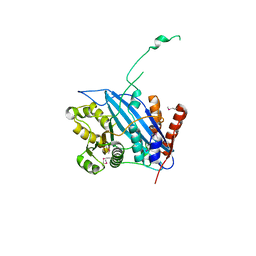 | | Crystal structure of Rtt109 | | Descriptor: | Regulator of Ty1 transposition protein 109 | | Authors: | Yuan, Y.A. | | Deposit date: | 2007-10-12 | | Release date: | 2008-09-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into histone h3 lysine 56 acetylation by rtt109
Structure, 16, 2008
|
|
8I0Q
 
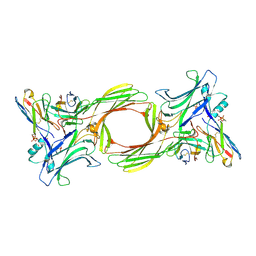 | | Structure of beta-arrestin1 in complex with a phosphopeptide corresponding to the human C-X-C chemokine receptor type 4, CXCR4 (Local refine) | | Descriptor: | Beta-arrestin-1, C-X-C chemokine receptor type 4, Fab30 Heavy Chain, ... | | Authors: | Maharana, J, Sarma, P, Yadav, M.K, Banerjee, R, Shukla, A.K. | | Deposit date: | 2023-01-11 | | Release date: | 2023-05-17 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.45 Å) | | Cite: | Structural snapshots uncover a key phosphorylation motif in GPCRs driving beta-arrestin activation.
Mol.Cell, 83, 2023
|
|
3SCF
 
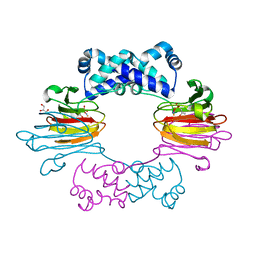 | | Fe(II)-HppE with S-HPP and NO | | Descriptor: | (S)-2-HYDROXYPROPYLPHOSPHONIC ACID, Epoxidase, FE (II) ION, ... | | Authors: | Drennan, C.L. | | Deposit date: | 2011-06-07 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural basis of regiospecificity of a mononuclear iron enzyme in antibiotic fosfomycin biosynthesis.
J.Am.Chem.Soc., 133, 2011
|
|
5HYN
 
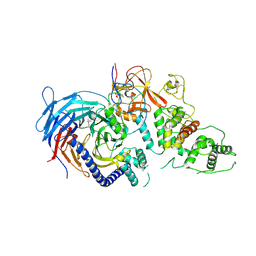 | | Structure of Human Polycomb Repressive Complex 2 (PRC2) with oncogenic histone H3K27M peptide | | Descriptor: | H3K27M, Histone-lysine N-methyltransferase EZH2, JARID2 K116me3, ... | | Authors: | Zhang, Y, Justin, N, Wilson, J.R, Gamblin, S.J. | | Deposit date: | 2016-02-01 | | Release date: | 2016-05-11 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural basis of oncogenic histone H3K27M inhibition of human polycomb repressive complex 2.
Nat Commun, 7, 2016
|
|
5ETA
 
 | | Structure of MAPK14 with bound the KIM domain of the Toxoplasma protein GRA24 | | Descriptor: | Mitogen-activated protein kinase 14, Putative transmembrane protein | | Authors: | Pellegrini, E, Palencia, A, Braun, L, Kapp, U, Bougdour, A, Belrhali, H, Bowler, M.W, Hakimi, M. | | Deposit date: | 2015-11-17 | | Release date: | 2016-10-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for the Subversion of MAP Kinase Signaling by an Intrinsically Disordered Parasite Secreted Agonist.
Structure, 25, 2017
|
|
8I3G
 
 | | Crystal structure of Eaf3-Eaf7 complex | | Descriptor: | Chromatin modification-related protein EAF3, Chromatin modification-related protein EAF7 | | Authors: | Chen, Z, Xu, C. | | Deposit date: | 2023-01-17 | | Release date: | 2023-05-31 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis for Eaf3-mediated assembly of Rpd3S and NuA4.
Cell Discov, 9, 2023
|
|
8I3F
 
 | |
8IIB
 
 | | Crystal structure of Israeli acute paralysis virus RNA-dependent RNA polymerase delta85 mutant (residues 86-546) | | Descriptor: | CADMIUM ION, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Fang, X, Lu, G, Hou, C, Gong, P. | | Deposit date: | 2023-02-24 | | Release date: | 2023-06-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Unusual substructure conformations observed in crystal structures of a dicistrovirus RNA-dependent RNA polymerase suggest contribution of the N-terminal extension in proper folding.
Virol Sin, 38, 2023
|
|
8IIC
 
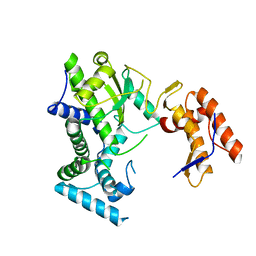 | |
9ESA
 
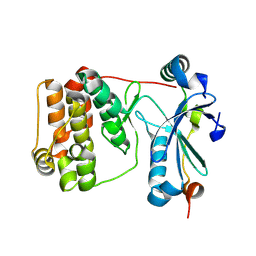 | | Aurora-C with SER mutation in complex with INCENP peptide | | Descriptor: | 1,2-ETHANEDIOL, Aurora kinase C, Inner centromere protein | | Authors: | Hillig, R.C. | | Deposit date: | 2024-03-26 | | Release date: | 2024-09-11 | | Last modified: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Surface-mutagenesis strategies to enable structural biology crystallization platforms.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8HST
 
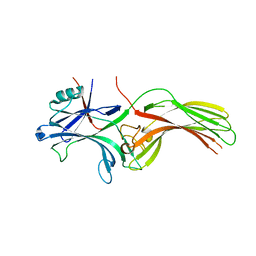 | |
8I60
 
 | |
8HSV
 
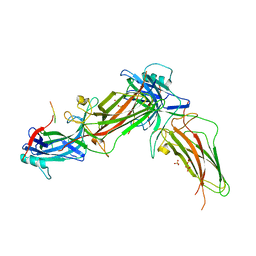 | | The structure of rat beta-arrestin1 in complex with a rat Mdm2 peptide | | Descriptor: | Beta-arrestin-1, SULFATE ION, peptide from E3 ubiquitin-protein ligase Mdm2 | | Authors: | Yun, Y, Yoon, H.J, Choi, Y, Lee, H.H. | | Deposit date: | 2022-12-20 | | Release date: | 2023-07-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | GPCR targeting of E3 ubiquitin ligase MDM2 by inactive beta-arrestin.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8IFF
 
 | | Cryo-EM structure of Arabidopsis phytochrome A. | | Descriptor: | 3-[5-[[(3~{R},4~{R})-3-ethyl-4-methyl-5-oxidanylidene-3,4-dihydropyrrol-2-yl]methyl]-2-[[5-[(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, Phytochrome A | | Authors: | Ma, L, Zhou, C, Wang, J, Guan, Z, Yin, P. | | Deposit date: | 2023-02-17 | | Release date: | 2023-08-02 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Plant phytochrome A in the Pr state assembles as an asymmetric dimer.
Cell Res., 33, 2023
|
|
9INR
 
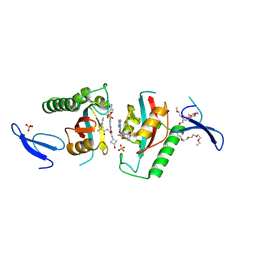 | | Crystal structure of PIN1 in complex with inhibitor C3 | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1, SULFATE ION, ... | | Authors: | Zhang, L.J, Zhang, L.Y. | | Deposit date: | 2024-07-08 | | Release date: | 2024-09-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Re-Evaluating PIN1 as a Therapeutic Target in Oncology Using Neutral Inhibitors and PROTACs.
J.Med.Chem., 67, 2024
|
|
9G40
 
 | |
2ZNY
 
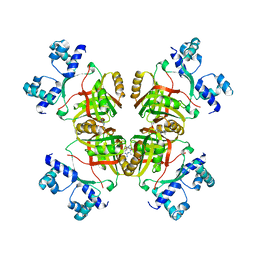 | | Crystal structure of the FFRP | | Descriptor: | ARGININE, Uncharacterized HTH-type transcriptional regulator PH1519 | | Authors: | Yamada, M, Suzuki, M. | | Deposit date: | 2008-05-02 | | Release date: | 2009-03-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Interactions between the archaeal transcription repressor FL11 and its coregulators lysine and arginine.
Proteins, 74, 2009
|
|
6X9B
 
 | |
6EBN
 
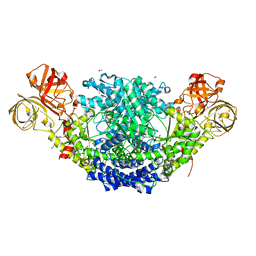 | | Crystal structure of Psilocybe cubensis noncanonical aromatic amino acid decarboxylase | | Descriptor: | FORMIC ACID, GLYCEROL, SODIUM ION, ... | | Authors: | Torrens-Spence, M.P, Chun-Ting, L, Pluskal, T, Chung, Y.K, Weng, J.K. | | Deposit date: | 2018-08-06 | | Release date: | 2018-12-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9663111 Å) | | Cite: | Monoamine Biosynthesis via a Noncanonical Calcium-Activatable Aromatic Amino Acid Decarboxylase in Psilocybin Mushroom.
ACS Chem. Biol., 13, 2018
|
|
9D7K
 
 | | Infectious B19V capsid | | Descriptor: | Alpha-1-antichymotrypsin | | Authors: | Lee, H, Hafenstein, S. | | Deposit date: | 2024-08-16 | | Release date: | 2024-10-16 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Infectious parvovirus B19 circulates in the blood coated with active host protease inhibitors.
Nat Commun, 15, 2024
|
|
1G5S
 
 | | CRYSTAL STRUCTURE OF HUMAN CYCLIN DEPENDENT KINASE 2 (CDK2) IN COMPLEX WITH THE INHIBITOR H717 | | Descriptor: | 2-[TRANS-(4-AMINOCYCLOHEXYL)AMINO]-6-(BENZYL-AMINO)-9-CYCLOPENTYLPURINE, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Dreyer, M.K, Borcherding, D.R, Dumont, J.A, Peet, N.P, Tsay, J.T, Wright, P.S, Bitonti, A.J, Shen, J, Kim, S.-H. | | Deposit date: | 2000-11-02 | | Release date: | 2001-11-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal structure of human cyclin-dependent kinase 2 in complex with the adenine-derived inhibitor H717.
J.Med.Chem., 44, 2001
|
|
9GCT
 
 | | Rho-ATP-Psu complex II expanded | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Polarity suppression protein, ... | | Authors: | Gjorgjevikj, D, Wahl, M.C, Hilal, T, Loll, B. | | Deposit date: | 2024-08-02 | | Release date: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Rho-ATP-Psu complex II expanded
To Be Published
|
|
6X9A
 
 | |
3NS9
 
 | | Crystal structure of CDK2 in complex with inhibitor BS-194 | | Descriptor: | (2S,3S)-3-{[7-(benzylamino)-3-(1-methylethyl)pyrazolo[1,5-a]pyrimidin-5-yl]amino}butane-1,2,4-triol, Cell division protein kinase 2 | | Authors: | Hazel, P, Freemont, P.S. | | Deposit date: | 2010-07-01 | | Release date: | 2010-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | A Novel Pyrazolo[1,5-a]pyrimidine Is a Potent Inhibitor of Cyclin-Dependent Protein Kinases 1, 2, and 9, Which Demonstrates Antitumor Effects in Human Tumor Xenografts Following Oral Administration.
J.Med.Chem., 53, 2010
|
|
