8QFO
 
 | | Ergothioneine dioxygenase, variant Y149F, from Thermocatellispora tengchongensis in complex with manganese | | Descriptor: | ACETATE ION, Cysteine dioxygenase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Vasseur, C.M, Seebeck, F.P. | | Deposit date: | 2023-09-04 | | Release date: | 2023-12-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Enzyme-Catalyzed Oxidative Degradation of Ergothioneine.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8QH6
 
 | | Crystal structure of IpgC in complex with a follow-up compound based on J20 | | Descriptor: | 3-azanyl-6-chloranyl-isoindol-1-one, CHLORIDE ION, Chaperone protein IpgC, ... | | Authors: | Wallbaum, J.E, Heine, A, Reuter, K. | | Deposit date: | 2023-09-06 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic Fragment Screening on the Shigella Type III Secretion System Chaperone IpgC.
Acs Omega, 8, 2023
|
|
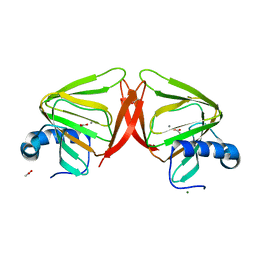
1B52
 
 | |
8QTG
 
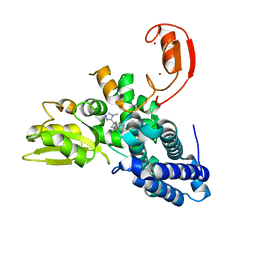 | | Crystal structure of CBL-b in complex with an allosteric inhibitor (compound 9) | | Descriptor: | 3-[3-[3-methyl-1-(4-methyl-1,2,4-triazol-3-yl)cyclobutyl]phenyl]-5-(trifluoromethyl)-1~{H}-pyridin-2-one, E3 ubiquitin-protein ligase CBL-B, SODIUM ION, ... | | Authors: | Schimpl, M. | | Deposit date: | 2023-10-12 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.419 Å) | | Cite: | Discovery, Optimization, and Biological Evaluation of Arylpyridones as Cbl-b Inhibitors.
J.Med.Chem., 67, 2024
|
|
8QTH
 
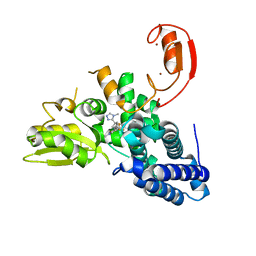 | | Crystal structure of CBL-b in complex with an allosteric inhibitor (compound 8) | | Descriptor: | 1-methyl-5-[3-[3-methyl-1-(4-methyl-1,2,4-triazol-3-yl)cyclobutyl]phenyl]-3-(trifluoromethyl)-7H-pyrrolo[2,3-b]pyridin-6-one, E3 ubiquitin-protein ligase CBL-B, SODIUM ION, ... | | Authors: | Schimpl, M. | | Deposit date: | 2023-10-12 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | Discovery, Optimization, and Biological Evaluation of Arylpyridones as Cbl-b Inhibitors.
J.Med.Chem., 67, 2024
|
|
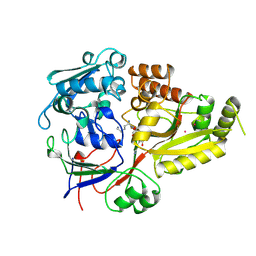
8RFH
 
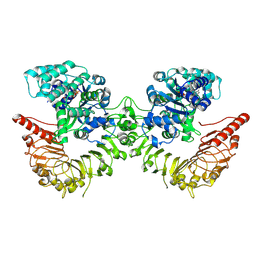 | |
8QM1
 
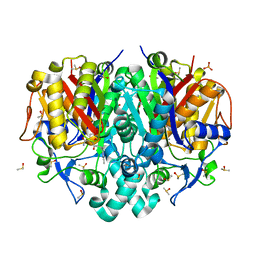 | | wild type Pa.FabF in complex cerulenin | | Descriptor: | (2S, 3R)-3-HYDROXY-4-OXO-7,10-TRANS,TRANS-DODECADIENAMIDE, 3-oxoacyl-[acyl-carrier-protein] synthase 2, ... | | Authors: | Georgiou, C, Brenk, R, Espeland, L.O. | | Deposit date: | 2023-09-21 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | New starting points for antibiotics targeting P. aeruginosa FabF discovered by crystallographic fragment screening followed by hit expansion
Chemrxiv, 2023
|
|
8R5I
 
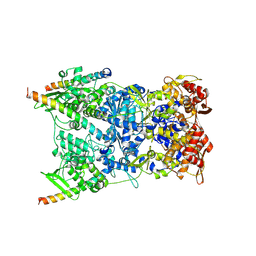 | | In situ structure of the Vaccinia virus (WR) A4/A10 palisade trimer in mature virions by flexible fitting into a cryoET map | | Descriptor: | Core protein A10, Core protein A4 | | Authors: | Calcraft, T, Hernandez-Gonzalez, M, Nans, A, Rosenthal, P.B, Way, M. | | Deposit date: | 2023-11-16 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (9.7 Å) | | Cite: | Palisade structure in intact vaccinia virions.
Mbio, 15, 2024
|
|
6XT2
 
 | | EQADH-NADH-HEPTAFLUOROBUTANOL, P21 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 2,2,3,3,4,4,4-heptafluorobutan-1-ol, ... | | Authors: | Plapp, B.V, Ramaswamy, S. | | Deposit date: | 2020-07-16 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Alternative binding modes in abortive NADH-alcohol complexes of horse liver alcohol dehydrogenase.
Arch.Biochem.Biophys., 701, 2021
|
|
8QTJ
 
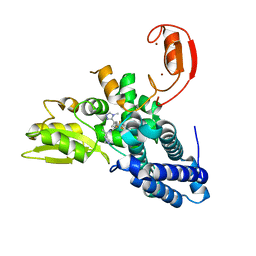 | | Crystal structure of Cbl-b in complex with an allosteric inhibitor (compound 30) | | Descriptor: | 3-[3-[3-methyl-1-(4-methyl-1,2,4-triazol-3-yl)cyclobutyl]phenyl]-1-[(1~{R})-1-(1-methylpyrazol-4-yl)ethyl]-5-(trifluoromethyl)pyridin-2-one, E3 ubiquitin-protein ligase CBL-B, SODIUM ION, ... | | Authors: | Schimpl, M. | | Deposit date: | 2023-10-12 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.523 Å) | | Cite: | Discovery, Optimization, and Biological Evaluation of Arylpyridones as Cbl-b Inhibitors.
J.Med.Chem., 67, 2024
|
|
8QMO
 
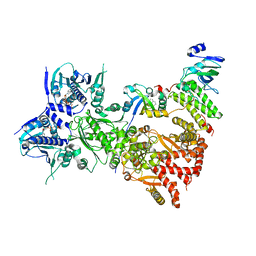 | | Cryo-EM structure of the benzo[a]pyrene-bound Hsp90-XAP2-AHR complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, AH receptor-interacting protein, Aryl hydrocarbon receptor, ... | | Authors: | Kwong, H.S, Grandvuillemin, L, Sirounian, S, Ancelin, A, Lai-Kee-Him, J, Carivenc, C, Lancey, C, Ragan, T.J, Hesketh, E.L, Bourguet, W, Gruszczyk, J. | | Deposit date: | 2023-09-24 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Structural Insights into the Activation of Human Aryl Hydrocarbon Receptor by the Environmental Contaminant Benzo[a]pyrene and Structurally Related Compounds.
J.Mol.Biol., 436, 2024
|
|
8QJR
 
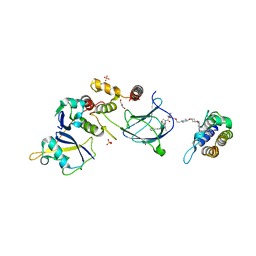 | | BRG1 bromodomain in complex with VBC via compound 17 | | Descriptor: | (2S,4R)-1-[(2R)-2-[3-[2-[4-[3-[4-[(1R,5S)-3-[3-azanyl-6-(2-hydroxyphenyl)pyridazin-4-yl]-3,8-diazabicyclo[3.2.1]octan-8-yl]pyridin-2-yl]oxycyclobutyl]oxypiperidin-1-yl]ethoxy]-1,2-oxazol-5-yl]-3-methyl-butanoyl]-N-[(1S)-1-[4-(4-methyl-1,3-thiazol-5-yl)phenyl]ethyl]-4-oxidanyl-pyrrolidine-2-carboxamide, CHLORIDE ION, Elongin-B, ... | | Authors: | Kerry, P.S, Hole, A.J, Perez-Dorado, J.I. | | Deposit date: | 2023-09-13 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | PROTACs Targeting BRM (SMARCA2) Afford Selective In Vivo Degradation over BRG1 (SMARCA4) and Are Active in BRG1 Mutant Xenograft Tumor Models.
J.Med.Chem., 67, 2024
|
|
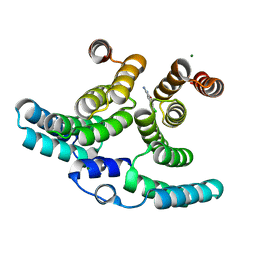
1B3L
 
 | |
8RI0
 
 | |
8R5C
 
 | | Crystal structure of human TRIM7 PRYSPRY domain bound to (2-(1-oxoisoindolin-2-yl)-3-phenylpropanoyl)-L-glutamine | | Descriptor: | (2~{S})-5-azanyl-5-oxidanylidene-2-[[(2~{S})-2-(3-oxidanylidene-1~{H}-isoindol-2-yl)-3-phenyl-propanoyl]amino]pentanoic acid, 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase TRIM7, ... | | Authors: | Munoz Sosa, C.J, Kraemer, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-11-16 | | Release date: | 2024-01-17 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A C-Degron Structure-Based Approach for the Development of Ligands Targeting the E3 Ligase TRIM7.
Acs Chem.Biol., 19, 2024
|
|
1B46
 
 | | OLIGO-PEPTIDE BINDING PROTEIN (OPPA) COMPLEXED WITH KPK | | Descriptor: | ACETATE ION, PROTEIN (LYS-PRO-LYS), PROTEIN (OLIGO-PEPTIDE BINDING PROTEIN), ... | | Authors: | Tame, J.R.H, Sleigh, S.H, Wilkinson, A.J. | | Deposit date: | 1999-01-05 | | Release date: | 1999-01-13 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic and calorimetric analysis of peptide binding to OppA protein.
J.Mol.Biol., 291, 1999
|
|
8QER
 
 | | Pseudomonas aeruginosa FabF C164A in complex with 4-(1H-pyrazole-3-carbonylamino)pentanoic acid | | Descriptor: | (4~{R})-4-(1~{H}-pyrazol-3-ylcarbonylamino)pentanoic acid, 1,2-ETHANEDIOL, 3-oxoacyl-[acyl-carrier-protein] synthase 2, ... | | Authors: | Yadrykhins'ky, V, Brenk, R, Georgiou, C. | | Deposit date: | 2023-09-01 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | New starting points for antibiotics targeting P. aeruginosa FabF discovered by crystallographic fragment screening followed by hit expansion
Chemrxiv, 2023
|
|
8QKY
 
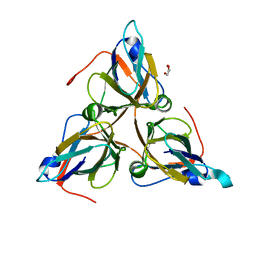 | | Bacteriophage T5 dUTPase | | Descriptor: | Deoxyuridine 5'-triphosphate nucleotidohydrolase, GLYCEROL | | Authors: | Gabdulkhakov, A.G, Dzhus, U.F, Selikhanov, G.K, Glukhov, A.S. | | Deposit date: | 2023-09-18 | | Release date: | 2024-01-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Bacteriophage T5 dUTPase: Combination of Common Enzymatic and Novel Functions.
Int J Mol Sci, 25, 2024
|
|
8QMX
 
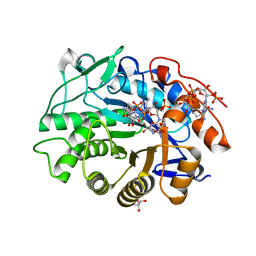 | | OPR3 wildtype in complex with NADPH4 | | Descriptor: | 12-oxophytodienoate reductase 3, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Bijelic, A, Macheroux, P, Kerschbaumer, B. | | Deposit date: | 2023-09-25 | | Release date: | 2024-01-17 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Loop 6 and the beta-hairpin flap are structural hotspots that determine cofactor specificity in the FMN-dependent family of ene-reductases.
Febs J., 291, 2024
|
|
1B47
 
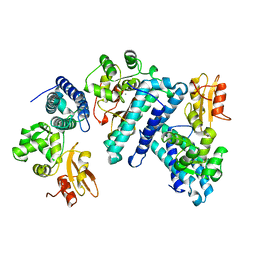 | | STRUCTURE OF THE N-TERMINAL DOMAIN OF CBL IN COMPLEX WITH ITS BINDING SITE IN ZAP-70 | | Descriptor: | CALCIUM ION, CBL | | Authors: | Meng, W, Sawasdikosol, S, Burakoff, S.J, Eck, M.J. | | Deposit date: | 1999-01-06 | | Release date: | 1999-04-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the amino-terminal domain of Cbl complexed to its binding site on ZAP-70 kinase.
Nature, 398, 1999
|
|
8QXB
 
 | | TDP-43 amyloid fibrils: Morphology-2 | | Descriptor: | TAR DNA-binding protein 43 | | Authors: | Sharma, K, Shenoy, J, Loquet, A, Schmidt, M, Faendrich, M. | | Deposit date: | 2023-10-24 | | Release date: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | Cryo-EM observation of the amyloid key structure of polymorphic TDP-43 amyloid fibrils.
Nat Commun, 15, 2024
|
|
8QXP
 
 | |
8R5D
 
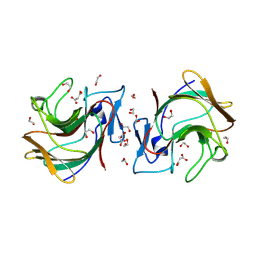 | | Crystal structure of human TRIM7 PRYSPRY domain | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase TRIM7, MALONIC ACID | | Authors: | Munoz Sosa, C.J, Kraemer, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-11-16 | | Release date: | 2024-01-17 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A C-Degron Structure-Based Approach for the Development of Ligands Targeting the E3 Ligase TRIM7.
Acs Chem.Biol., 19, 2024
|
|
8RNW
 
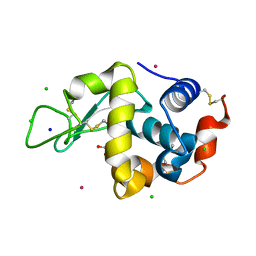 | | Hen Egg White Lysozyme soaked with trans-Ru(DMSO)4Cl2 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Oszajca, M, Flejszar, M, Szura, A, Drozdz, P, Brindell, M, Kurpiewska, K. | | Deposit date: | 2024-01-11 | | Release date: | 2024-01-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Exploring the coordination chemistry of ruthenium complexes with lysozymes: structural and in-solution studies.
Front Chem, 12, 2024
|
|
8QLD
 
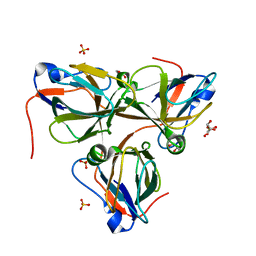 | | Bacteriophage T5 dUTPase mutant with loop deletion (30-35 aa) | | Descriptor: | CHLORIDE ION, Deoxyuridine 5'-triphosphate nucleotidohydrolase, GLYCEROL, ... | | Authors: | Gabdulkhakov, A.G, Dzhus, U.F, Selikhanov, G.K, Glukhov, A.S. | | Deposit date: | 2023-09-19 | | Release date: | 2024-01-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Bacteriophage T5 dUTPase: Combination of Common Enzymatic and Novel Functions.
Int J Mol Sci, 25, 2024
|
|
