8JSP
 
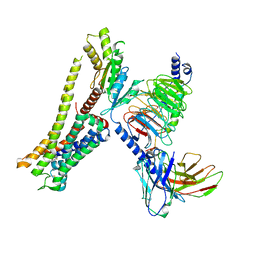 | | Ulotaront(SEP-363856)-bound Serotonin 1A (5-HT1A) receptor-Gi complex | | Descriptor: | 1-[(7~{S})-5,7-dihydro-4~{H}-thieno[2,3-c]pyran-7-yl]-~{N}-methyl-methanamine, 5-hydroxytryptamine receptor 1A, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Xu, Z, Guo, L.L, Zhao, C, Shen, S.Y, Sun, J.P, Shao, Z.H. | | Deposit date: | 2023-06-20 | | Release date: | 2023-11-15 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Ligand recognition and G-protein coupling of trace amine receptor TAAR1.
Nature, 624, 2023
|
|
5UGM
 
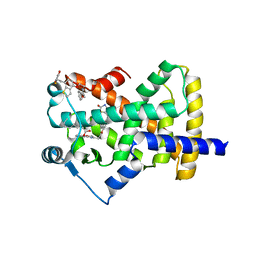 | | Crystal Structure of Human PPARgamma Ligand Binding Domain in Complex with Edaglitazone | | Descriptor: | (5R)-5-({4-[2-(5-methyl-2-phenyl-1,3-oxazol-4-yl)ethoxy]-1-benzothiophen-7-yl}methyl)-1,3-thiazolidine-2,4-dione, Peroxisome proliferator-activated receptor gamma, nonanoic acid | | Authors: | Shang, J, Kojetin, D.J. | | Deposit date: | 2017-01-09 | | Release date: | 2018-01-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Cooperative cobinding of synthetic and natural ligands to the nuclear receptor PPAR gamma.
Elife, 7, 2018
|
|
4TY9
 
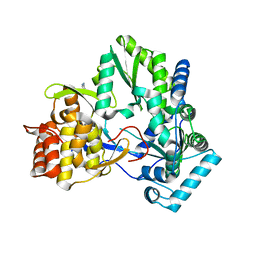 | | An Ligand-observed Mass Spectrometry-based Approach Integrated into the Fragment Based Lead Discovery Pipeline | | Descriptor: | 5-(trifluoromethyl)pyridin-2-amine, Polyprotein | | Authors: | Shui, W, Yang, C, Lin, J, Chen, X, Qin, S, Chen, S. | | Deposit date: | 2014-07-08 | | Release date: | 2015-05-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | A ligand-observed mass spectrometry approach integrated into the fragment based lead discovery pipeline
Sci Rep, 5, 2015
|
|
1CCJ
 
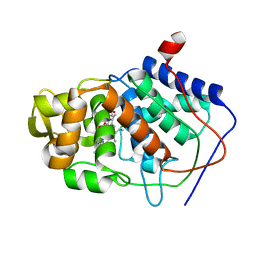 | | CONFORMER SELECTION BY LIGAND BINDING OBSERVED WITH PROTEIN CRYSTALLOGRAPHY | | Descriptor: | CYTOCHROME C PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Cao, Y, Musah, R.A, Wilcox, S.K, Goodin, D.B, Mcree, D.E. | | Deposit date: | 1997-01-22 | | Release date: | 1997-07-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Protein conformer selection by ligand binding observed with crystallography.
Protein Sci., 7, 1998
|
|
4TYB
 
 | | An Ligand-observed Mass Spectrometry-based Approach Integrated into the Fragment Based Lead Discovery Pipeline | | Descriptor: | (2R)-morpholin-4-yl(phenyl)ethanenitrile, Polyprotein | | Authors: | Shui, W, Yang, C, Lin, J, Chen, X, Qin, S, Chen, S. | | Deposit date: | 2014-07-08 | | Release date: | 2015-05-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | A ligand-observed mass spectrometry approach integrated into the fragment based lead discovery pipeline
Sci Rep, 5, 2015
|
|
4TYA
 
 | | An Ligand-observed Mass Spectrometry-based Approach Integrated into the Fragment Based Lead Discovery Pipeline | | Descriptor: | 4-(trifluoromethyl)benzoic acid, Polyprotein | | Authors: | Shui, W, Yang, C, Lin, J, Chen, X, Qin, S, Chen, S. | | Deposit date: | 2014-07-08 | | Release date: | 2015-05-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | A ligand-observed mass spectrometry approach integrated into the fragment based lead discovery pipeline
Sci Rep, 5, 2015
|
|
4TXS
 
 | | An Ligand-observed Mass Spectrometry-based Approach Integrated into the Fragment Based Lead Discovery Pipeline | | Descriptor: | (4-hydroxyphenyl)acetonitrile, Polyprotein | | Authors: | Shui, W, Yang, C, Lin, J, Chen, X, Qin, S, Chen, S. | | Deposit date: | 2014-07-07 | | Release date: | 2015-05-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | A ligand-observed mass spectrometry approach integrated into the fragment based lead discovery pipeline
Sci Rep, 5, 2015
|
|
5DGW
 
 | | Crystal Structure of HIV-1 Protease Inhibitor GRL-105-11A Containing Substituted fused-Tetrahydropyranyl Tetrahydrofuran as P2-Ligand | | Descriptor: | (3R,3aS,4S,7aS)-3-(ethylamino)hexahydro-4H-furo[2,3-b]pyran-4-yl [(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-phenylbutan-2-yl]carbamate, CHLORIDE ION, Pol protein, ... | | Authors: | Agniswamy, J, Wang, Y.-F, Weber, I.T. | | Deposit date: | 2015-08-28 | | Release date: | 2015-10-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Design, synthesis, biological evaluation and X-ray structural studies of HIV-1 protease inhibitors containing substituted fused-tetrahydropyranyl tetrahydrofuran as P2-ligands.
Org.Biomol.Chem., 13, 2015
|
|
2AX6
 
 | | Crystal Structure Of The Androgen Receptor Ligand Binding Domain T877A Mutant In Complex With Hydroxyflutamide | | Descriptor: | Androgen receptor, HYDROXYFLUTAMIDE | | Authors: | Bohl, C.E, Miller, D.D, Chen, J, Bell, C.E, Dalton, J.T. | | Deposit date: | 2005-09-03 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis for Accommodation of Nonsteroidal Ligands in the Androgen Receptor
J.Biol.Chem., 280, 2005
|
|
2AXA
 
 | | Crystal Structure Of The Androgen Receptor Ligand Binding Domain In Complex With S-1 | | Descriptor: | Androgen receptor, S-3-(4-FLUOROPHENOXY)-2-HYDROXY-2-METHYL-N-[4-NITRO-3-(TRIFLUOROMETHYL)PHENYL]PROPANAMIDE | | Authors: | Bohl, C.E, Miller, D.D, Chen, J, Bell, C.E, Dalton, J.T. | | Deposit date: | 2005-09-03 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for Accommodation of Nonsteroidal Ligands in the Androgen Receptor
J.Biol.Chem., 280, 2005
|
|
2AX7
 
 | | Crystal Structure Of The Androgen Receptor Ligand Binding Domain T877A Mutant In Complex With S-1 | | Descriptor: | Androgen receptor, S-3-(4-FLUOROPHENOXY)-2-HYDROXY-2-METHYL-N-[4-NITRO-3-(TRIFLUOROMETHYL)PHENYL]PROPANAMIDE | | Authors: | Bohl, C.E, Miller, D.D, Chen, J, Bell, C.E, Dalton, J.T. | | Deposit date: | 2005-09-03 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for Accommodation of Nonsteroidal Ligands in the Androgen Receptor
J.Biol.Chem., 280, 2005
|
|
6PKF
 
 | | Myocilin OLF mutant N428E/D478K | | Descriptor: | GLYCEROL, Myocilin | | Authors: | Lieberman, R.L, Hill, S.E. | | Deposit date: | 2019-06-29 | | Release date: | 2019-09-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.484 Å) | | Cite: | Calcium-ligand variants of the myocilin olfactomedin propeller selected from invertebrate phyla reveal cross-talk with N-terminal blade and surface helices.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
1JW4
 
 | | Structure of ligand-free maltodextrin-binding protein | | Descriptor: | maltodextrin-binding protein | | Authors: | Duan, X, Quiocho, F.A. | | Deposit date: | 2001-09-02 | | Release date: | 2002-01-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural evidence for a dominant role of nonpolar interactions in the binding of a transport/chemosensory receptor to its highly polar ligands.
Biochemistry, 41, 2002
|
|
2R5W
 
 | | Crystal structure of a bifunctional NMN adenylyltransferase/ADP ribose pyrophosphatase from Francisella tularensis | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Nicotinamide-nucleotide adenylyltransferase | | Authors: | Huang, N, Sorci, L, Zhang, X, Brautigan, C, Li, X, Raffaelli, N, Grishin, N, Osterman, A, Zhang, H. | | Deposit date: | 2007-09-04 | | Release date: | 2008-03-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Bifunctional NMN Adenylyltransferase/ADP-Ribose Pyrophosphatase: Structure and Function in Bacterial NAD Metabolism.
Structure, 16, 2008
|
|
6PKD
 
 | | Myocilin OLF mutant N428D/D478H | | Descriptor: | Myocilin, SODIUM ION | | Authors: | Lieberman, R.L, Hill, S.E. | | Deposit date: | 2019-06-29 | | Release date: | 2019-09-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Calcium-ligand variants of the myocilin olfactomedin propeller selected from invertebrate phyla reveal cross-talk with N-terminal blade and surface helices.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
2JU7
 
 | | Solution-State Structures of Oleate-Liganded LFABP, Protein Only | | Descriptor: | Fatty acid-binding protein, liver | | Authors: | He, Y, Yang, X, Wang, H, Estephan, R, Francis, F, Kodukula, S, Storch, J, Stark, R.E. | | Deposit date: | 2007-08-15 | | Release date: | 2007-11-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution-State Molecular Structure of Apo and Oleate-Liganded Liver Fatty Acid-Binding Protein
Biochemistry, 46, 2007
|
|
5MKU
 
 | | Crystal structure of the Retinoid X Receptor alpha in complex with synthetic honokiol derivative 4 and a fragment of the TIF2 co-activator. | | Descriptor: | (~{E})-3-[4-oxidanyl-3-(3-propan-2-ylphenyl)phenyl]prop-2-enoic acid, HIS-LYS-ILE-LEU-HIS-ARG-LEU-LEU-GLN-ASP-SER, Retinoic acid receptor RXR-alpha | | Authors: | Andrei, S.A, Brunsveld, L, Scheepstra, M, Ottmann, C. | | Deposit date: | 2016-12-05 | | Release date: | 2017-11-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Ligand Dependent Switch from RXR Homo- to RXR-NURR1 Heterodimerization.
ACS Chem Neurosci, 8, 2017
|
|
4YT1
 
 | | Human PPAR Gamma Ligand Binding Domain in complex with a Gammma Selective Synthetic Partial Agonist MEKT76 | | Descriptor: | N-(benzylsulfonyl)-4-propoxy-3-({[4-(pyrimidin-2-yl)benzoyl]amino}methyl)benzamide, Peroxisome proliferator-activated receptor gamma | | Authors: | Oyama, T, Ohashi, M, Miyachi, H, Kusunoki, M. | | Deposit date: | 2015-03-17 | | Release date: | 2016-03-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Peroxisome proliferator-activated receptor gamma (PPAR gamma ) has multiple binding points that accommodate ligands in various conformations: Structurally similar PPAR gamma partial agonists bind to PPAR gamma LBD in different conformations
Bioorg.Med.Chem.Lett., 25, 2015
|
|
6E0A
 
 | | Crystal Structure of Helicobacter pylori TlpA Chemoreceptor Ligand Binding Domain | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Remington, S.J, Guillemin, K, Sweeney, E, Perkins, A. | | Deposit date: | 2018-07-06 | | Release date: | 2018-09-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structures of the ligand-binding domain of Helicobacter pylori chemoreceptor TlpA.
Protein Sci., 27, 2018
|
|
5M39
 
 | | Crystal structure of BRD4 BROMODOMAIN 1 IN COMPLEX WITH LIGAND 1 | | Descriptor: | 6-(3,4-dimethoxyphenyl)-3-methyl-[1,2,4]triazolo[4,3-b]pyridazine, Bromodomain-containing protein 4 | | Authors: | Kessler, D, Mayer, M, Engelhardt, H, Wolkerstorfer, B, Geist, L. | | Deposit date: | 2016-10-14 | | Release date: | 2017-09-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Direct NMR Probing of Hydration Shells of Protein Ligand Interfaces and Its Application to Drug Design.
J. Med. Chem., 60, 2017
|
|
2BU7
 
 | | crystal structures of human pyruvate dehydrogenase kinase 2 containing physiological and synthetic ligands | | Descriptor: | N-(2-AMINOETHYL)-2-{3-CHLORO-4-[(4-ISOPROPYLBENZYL)OXY]PHENYL} ACETAMIDE, PYRUVATE DEHYDROGENASE KINASE ISOENZYME 2 | | Authors: | Knoechel, T.R, Tucker, A.D, Robinson, C.M, Phillips, C, Taylor, W, Bungay, P.J, Kasten, S.A, Roche, T.E, Brown, D.G. | | Deposit date: | 2005-06-08 | | Release date: | 2006-02-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Regulatory Roles of the N-Terminal Domain Based on Crystal Structures of Human Pyruvate Dehydrogenase Kinase 2 Containing Physiological and Synthetic Ligands.
Biochemistry, 45, 2006
|
|
1MS7
 
 | | X-ray structure of the GluR2 ligand-binding core (S1S2J) in complex with (S)-Des-Me-AMPA at 1.97 A resolution, Crystallization in the presence of zinc acetate | | Descriptor: | (S)-2-AMINO-3-(3-HYDROXY-ISOXAZOL-4-YL)PROPIONIC ACID, Glutamate receptor subunit 2, ZINC ION | | Authors: | Kasper, C, Lunn, M.-L, Liljefors, T, Gouaux, E, Egebjerg, J, Kastrup, J.S. | | Deposit date: | 2002-09-19 | | Release date: | 2003-07-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | GluR2 ligand-binding core complexes: importance of the isoxazolol moiety and 5-substituent for the binding mode of AMPA-type agonists
FEBS Lett., 531, 2002
|
|
3RLH
 
 | | Crystal structure of a class II phospholipase D from Loxosceles intermedia venom | | Descriptor: | (2r,5r)-5-amino-2-hydroxy-5-(hydroxymethyl)-1,3,2lambda~5~-dioxaphosphinan-2-one, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Giuseppe, P.O, Ullah, A, Veiga, S.S, Murakami, M.T, Arni, R.K, Doherty, D.Z, Gismene, C, Bachega, J.F.R, Chahine, J, Gonzalez, J.E.H. | | Deposit date: | 2011-04-19 | | Release date: | 2011-06-29 | | Last modified: | 2025-02-19 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structure of a novel class II phospholipase D: Catalytic cleft is modified by a disulphide bridge.
Biochem.Biophys.Res.Commun., 409, 2011
|
|
3KD7
 
 | |
4Q02
 
 | | Second-site screening of K-Ras in the presence of covalently attached first-site ligands | | Descriptor: | 3,4-difluorobenzenethiol, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Sun, Q, Phan, J, Friberg, A, Camper, D.V, Olejniczak, E.T, Fesik, S.W. | | Deposit date: | 2014-03-31 | | Release date: | 2014-09-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | A method for the second-site screening of K-Ras in the presence of a covalently attached first-site ligand.
J.Biomol.Nmr, 60, 2014
|
|
