1O9F
 
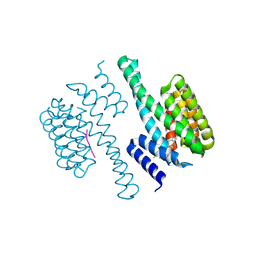 | | Structural view of a fungal toxin acting on a 14-3-3 regulatory complex | | Descriptor: | 14-3-3-LIKE PROTEIN C, FUSICOCCIN, PLASMA MEMBRANE H+ ATPASE | | Authors: | Wurtele, M, Jelich-Ottmann, C, Wittinghofer, A, Oecking, C. | | Deposit date: | 2002-12-12 | | Release date: | 2003-03-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural View of a Fungal Toxin Acting on a 14-3-3 Regulatory Complex
Embo J., 22, 2003
|
|
1PAF
 
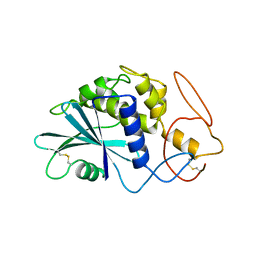 | | THE 2.5 ANGSTROMS STRUCTURE OF POKEWEED ANTIVIRAL PROTEIN | | Descriptor: | POKEWEED ANTIVIRAL PROTEIN | | Authors: | Monzingo, A.F, Collins, E.J, Ernst, S.R, Irvin, J.D, Robertus, J.D. | | Deposit date: | 1992-10-19 | | Release date: | 1994-01-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The 2.5 A structure of pokeweed antiviral protein.
J.Mol.Biol., 233, 1993
|
|
1P8C
 
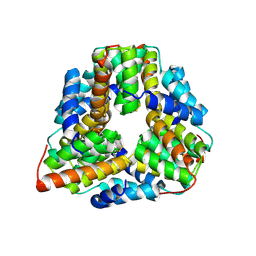 | | Crystal structure of TM1620 (APC4843) from Thermotoga maritima | | Descriptor: | conserved hypothetical protein | | Authors: | Kim, Y, Joachimiak, A, Brunzelle, J.S, Korolev, S.V, Edwards, A, Xu, X, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-05-06 | | Release date: | 2003-09-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure Analysis of Thermotoga maritima protein TM1620 (APC4843)
To be Published
|
|
1OY1
 
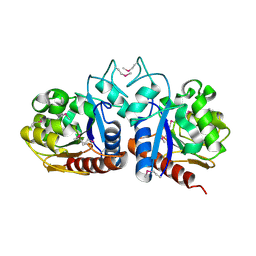 | | X-Ray Structure Of ElbB From E. Coli. Northeast Structural Genomics Research Consortium (Nesg) Target Er105 | | Descriptor: | PUTATIVE sigma cross-reacting protein 27A | | Authors: | Benach, J, Edstrom, W, Ma, L.C, Xiao, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-04-03 | | Release date: | 2003-04-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | X-Ray Structure Of ElbB From E. Coli. Northeast Structural Genomics Research Consortium (Nesg) Target Er105
To be Published
|
|
1OYQ
 
 | | TRYPSIN INHIBITOR COMPLEX | | Descriptor: | CALCIUM ION, SULFATE ION, Trypsin, ... | | Authors: | Nar, H. | | Deposit date: | 2003-04-07 | | Release date: | 2003-04-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for Inhibition Promiscuity of Dual Specific Thrombin and Factor Xa Blood Coagulation Inhibitors
Structure, 9, 2001
|
|
1P0C
 
 | | Crystal Structure of the NADP(H)-Dependent Vertebrate Alcohol Dehydrogenase (ADH8) | | Descriptor: | GLYCEROL, NADP-dependent ALCOHOL DEHYDROGENASE, PHOSPHATE ION, ... | | Authors: | Rosell, A, Valencia, E, Pares, X, Fita, I, Farres, J, Ochoa, W.F. | | Deposit date: | 2003-04-10 | | Release date: | 2003-04-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Vertebrate NADP(H)-dependent Alcohol Dehydrogenase (ADH8)
J.Mol.Biol., 330, 2003
|
|
1P57
 
 | | Extracellular domain of human hepsin | | Descriptor: | 2-{5-[AMINO(IMINIO)METHYL]-1H-BENZIMIDAZOL-2-YL}BENZENOLATE, Serine protease hepsin | | Authors: | Somoza, J.R, Ho, J.D, Luong, C, Sprengeler, P.A, Mortara, K, Shrader, W.D, Sperandio, D, Chan, H, McGrath, M.E, Katz, B.A. | | Deposit date: | 2003-04-25 | | Release date: | 2004-01-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The Structure of the Extracellular Region of Human Hepsin Reveals a Serine Protease Domain and a Novel Scavenger Receptor Cysteine-Rich (SRCR) Domain
Structure, 11, 2003
|
|
1OTJ
 
 | | Crystal structure of APO (iron-free) TauD | | Descriptor: | 2-AMINOETHANESULFONIC ACID, Alpha-Ketoglutarate-Dependent Taurine Dioxygenase, CHLORIDE ION | | Authors: | O'Brien, J.R, Schuller, D.J, Yang, V.S, Dillard, B.D, Lanzilotta, W.N. | | Deposit date: | 2003-03-21 | | Release date: | 2003-09-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Substrate-induced conformational changes in Escherichia coli taurine/alpha-ketoglutarate dioxygenase and insight into the oligomeric structure
Biochemistry, 42, 2003
|
|
4GD3
 
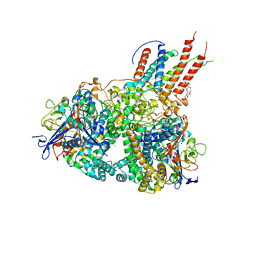 | | Structure of E. coli hydrogenase-1 in complex with cytochrome b | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Volbeda, A, Fontecilla-Camps, J.C, Darnault, C. | | Deposit date: | 2012-07-31 | | Release date: | 2013-01-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal Structure of the O(2)-Tolerant Membrane-Bound Hydrogenase 1 from Escherichia coli in Complex with Its Cognate Cytochrome b.
Structure, 21, 2013
|
|
1OWG
 
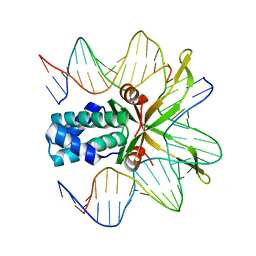 | | Crystal structure of WT IHF complexed with an altered H' site (T44A) | | Descriptor: | 5'-D(*GP*CP*TP*TP*AP*TP*CP*AP*AP*TP*TP*TP*GP*TP*AP*GP*CP*AP*CP*C)-3', 5'-D(*GP*GP*CP*CP*AP*AP*AP*AP*AP*AP*GP*CP*AP*TP*T)-3', Integration Host Factor Alpha-subunit, ... | | Authors: | Lynch, T.W, Read, E.K, Mattis, A.N, Gardner, J.F, Rice, P.A. | | Deposit date: | 2003-03-28 | | Release date: | 2003-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Integration Host Factor: putting a twist on protein-DNA recognition
J.Mol.Biol., 330, 2003
|
|
1OPE
 
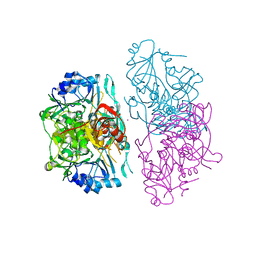 | | Deletion mutant of SUCCINYL-COA:3-KETOACID COA TRANSFERASE FROM PIG HEART | | Descriptor: | MERCURY (II) ION, POTASSIUM ION, Succinyl-CoA:3-ketoacid-coenzyme A transferase | | Authors: | Coros, A.M, Swenson, L, Wolodko, W.T, Fraser, M.E. | | Deposit date: | 2003-03-05 | | Release date: | 2004-04-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the CoA transferase from pig heart to 1.7 A resolution.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1OQ1
 
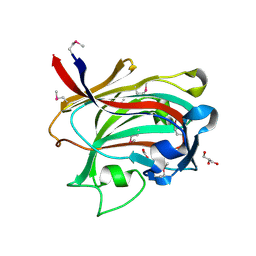 | |
1OWJ
 
 | | Substituted 2-Naphthamidine Inhibitors of Urokinase | | Descriptor: | 6-[(Z)-AMINO(IMINO)METHYL]-N-(1-ISOPROPYL-3,4-DIHYDROISOQUINOLIN-7-YL)-2-NAPHTHAMIDE, Urokinase-type plasminogen activator | | Authors: | Wendt, M.D, Rockway, T.W, Geyer, A, McClellan, W, Weitzberg, M, Zhao, X, Mantei, R, Nienaber, V.L, Stewart, K, Klinghofer, V, Giranda, V.L. | | Deposit date: | 2003-03-28 | | Release date: | 2003-09-30 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Identification of Novel Binding Interactions in the Development of Potent, Selective 2-Naphthamidine Inhibitors of Urokinase. Synthesis, Structural Analysis, and SAR of N-Phenyl Amide 6-Substitution.
J.Med.Chem., 47, 2004
|
|
4PM1
 
 | | Human transthyretin (TTR) complexed with 16-alpha-bromo-estradiol | | Descriptor: | (14beta,16alpha,17alpha)-16-bromoestra-1,3,5(10)-triene-3,17-diol, 1,2-ETHANEDIOL, Transthyretin | | Authors: | Stura, E.A, Ciccone, L. | | Deposit date: | 2014-05-20 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Transthyretin complexes with curcumin and bromo-estradiol: evaluation of solubilizing multicomponent mixtures.
N Biotechnol, 32, 2014
|
|
1OTP
 
 | | STRUCTURAL AND THEORETICAL STUDIES SUGGEST DOMAIN MOVEMENT PRODUCES AN ACTIVE CONFORMATION OF THYMIDINE PHOSPHORYLASE | | Descriptor: | THYMIDINE PHOSPHORYLASE | | Authors: | Pugmire, M.J, Cook, W.J, Jasanoff, A, Walter, M.R, Ealick, S.E. | | Deposit date: | 1997-11-09 | | Release date: | 1998-12-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and theoretical studies suggest domain movement produces an active conformation of thymidine phosphorylase.
J.Mol.Biol., 281, 1998
|
|
1OSZ
 
 | | MHC CLASS I H-2KB HEAVY CHAIN COMPLEXED WITH BETA-2 MICROGLOBULIN AND AN (L4V) MUTANT OF THE VESICULAR STOMATITIS VIRUS NUCLEOPROTEIN | | Descriptor: | BETA-2 MICROGLOBULIN, MHC CLASS I H-2KB HEAVY CHAIN, VESICULAR STOMATITIS VIRUS NUCLEOPROTEIN | | Authors: | Ghendler, Y, Teng, M.-K, Liu, J.-H, Witte, T, Liu, J, Kim, K.S, Kern, P, Chang, H.-C, Wang, J.-H, Reinherz, E.L. | | Deposit date: | 1998-06-18 | | Release date: | 1999-01-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Differential thymic selection outcomes stimulated by focal structural alteration in peptide/major histocompatibility complex ligands.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1OT5
 
 | | The 2.4 Angstrom Crystal Structure of Kex2 in complex with a peptidyl-boronic acid inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ac-Ala-Lys-boroArg N-acetylated boronic acid peptide inhibitor, ... | | Authors: | Holyoak, T, Wilson, M.A, Fenn, T.D, Kettner, C.A, Petsko, G.A, Fuller, R.S, Ringe, D. | | Deposit date: | 2003-03-21 | | Release date: | 2003-06-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 2.4 A Resolution Crystal Structure of the Prototypical Hormone-Processing Protease Kex2 in Complex with an Ala-Lys-Arg Boronic Acid Inhibitor
Biochemistry, 42, 2003
|
|
1P14
 
 | | Crystal structure of a catalytic-loop mutant of the insulin receptor tyrosine kinase | | Descriptor: | insulin receptor | | Authors: | Li, S, Covino, N.D, Stein, E.G, Till, J.H, Hubbard, S.R. | | Deposit date: | 2003-04-11 | | Release date: | 2003-07-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and biochemical evidence for an autoinhibitory role for tyrosine 984 in the juxtamembrane region of the insulin receptor
J.Biol.Chem., 278, 2003
|
|
1P2S
 
 | | H-Ras 166 in 50% 2,2,2 triflouroethanol | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, TRIFLUOROETHANOL, ... | | Authors: | Buhrman, G.K, de Serrano, V, Mattos, C. | | Deposit date: | 2003-04-16 | | Release date: | 2003-08-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Organic Solvents Order the Dynamic Switch II in Ras Crystals
Structure, 11, 2003
|
|
1OWK
 
 | | Substituted 2-Naphthamidine Inhibitors of Urokinase | | Descriptor: | 6-[(Z)-AMINO(IMINO)METHYL]-N-(1-ISOPROPYL-1,2,3,4-TETRAHYDROISOQUINOLIN-7-YL)-2-NAPHTHAMIDE, Urokinase-type plasminogen activator | | Authors: | Wendt, M.D, Rockway, T.W, Geyer, A, McClellan, W, Weitzberg, M, Zhao, X, Mantei, R, Nienaber, V.L, Stewart, K, Klinghofer, V, Giranda, V.L. | | Deposit date: | 2003-03-28 | | Release date: | 2003-09-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Identification of Novel Binding Interactions in the Development of Potent, Selective 2-Naphthamidine Inhibitors of Urokinase. Synthesis, Structural Analysis, and SAR of N-Phenyl Amide 6-Substitution.
J.Med.Chem., 47, 2004
|
|
1P8F
 
 | |
1OX5
 
 | |
1OZY
 
 | |
1OM4
 
 | | STRUCTURE OF RAT NEURONAL NOS HEME DOMAIN WITH L-ARGININE BOUND | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, ARGININE, ... | | Authors: | Li, H, Martasek, P, Shimizu, H, Masters, B.S.S, Poulos, T.L, Raman, C.S. | | Deposit date: | 2003-02-24 | | Release date: | 2003-03-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of Rat Neuronal NOS Heme Domain with L-Arginine Bound
To be Published
|
|
1OMP
 
 | |
