1N1I
 
 | | The structure of MSP-1(19) from Plasmodium knowlesi | | Descriptor: | HISTIDINE, IMIDAZOLE, Merozoite surface protein-1 | | Authors: | Garman, S.C, Simcoke, W.N, Stowers, A.W, Garboczi, D.N. | | Deposit date: | 2002-10-17 | | Release date: | 2003-02-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the C-terminal domains of merozoite surface protein-1 from
Plasmodium knowlesi reveals a novel histidine binding site
J.Biol.Chem., 278, 2003
|
|
3KYF
 
 | | Crystal structure of P4397 complexed with c-di-GMP | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, Putative uncharacterized protein | | Authors: | Ryu, K.S, Ko, J, Kim, H, Choi, B.S. | | Deposit date: | 2009-12-06 | | Release date: | 2010-04-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of PP4397 Reveals the Molecular Basis for Different c-di-GMP Binding Modes by Pilz Domain Proteins.
J.Mol.Biol., 398, 2010
|
|
2DJF
 
 | | Crystal Structure of human dipeptidyl peptidase I (Cathepsin C) in complex with the inhibitor Gly-Phe-CHN2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETIC ACID, CHLORIDE ION, ... | | Authors: | Molgaard, A, Arnau, J, Lauritzen, C, Larsen, S, Petersen, G, Pedersen, J. | | Deposit date: | 2006-04-02 | | Release date: | 2006-11-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of human dipeptidyl peptidase I (cathepsin C) in complex with the inhibitor Gly-Phe-CHN2
Biochem.J., 401, 2007
|
|
4EI4
 
 | | JAK1 kinase (JH1 domain) in complex with compound 20 | | Descriptor: | (1R,3R)-3-(2-methylimidazo[4,5-d]pyrrolo[2,3-b]pyridin-1(8H)-yl)cyclohexanol, Tyrosine-protein kinase JAK1 | | Authors: | Eigenbrot, C, Steffek, M. | | Deposit date: | 2012-04-04 | | Release date: | 2012-07-04 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Discovery and optimization of C-2 methyl imidazopyrrolopyridines as potent and orally bioavailable JAK1 inhibitors with selectivity over JAK2.
J.Med.Chem., 55, 2012
|
|
2AJE
 
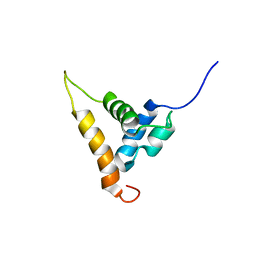 | | Solution structure of the Arabidopsis thaliana telomeric repeat-binding protein DNA binding domain | | Descriptor: | telomere repeat-binding protein | | Authors: | Hsiao, H.H, Sue, S.C, Chung, B.C, Cheng, Y.H, Huang, T.H. | | Deposit date: | 2005-08-01 | | Release date: | 2006-07-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Arabidopsis thaliana telomeric repeat-binding protein DNA binding domain: a new fold with an additional C-terminal helix
J.Mol.Biol., 356, 2006
|
|
4QDS
 
 | | Physical basis for Nrp2 ligand binding | | Descriptor: | ACETATE ION, GLYCEROL, Neuropilin-2 | | Authors: | Parker, M.W, Vander Kooi, C.W. | | Deposit date: | 2014-05-14 | | Release date: | 2015-04-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis for VEGF-C Binding to Neuropilin-2 and Sequestration by a Soluble Splice Form.
Structure, 23, 2015
|
|
1I8O
 
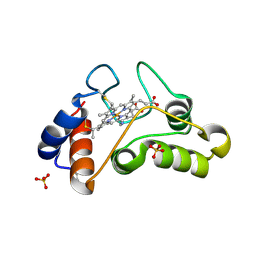 | |
4F08
 
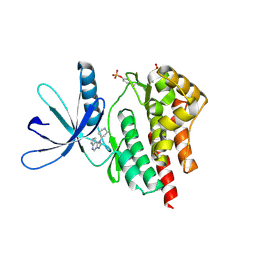 | |
7CJX
 
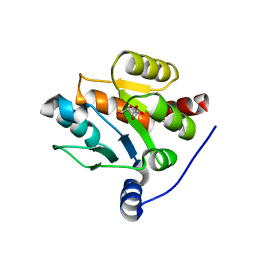 | | UDP-glucuronosyltransferase 2B15 C-terminal domain-L446S | | Descriptor: | L(+)-TARTARIC ACID, UDP-glucuronosyltransferase 2B15 | | Authors: | Wang, C.Y, Zhang, L. | | Deposit date: | 2020-07-14 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.986414 Å) | | Cite: | Structure of UDP-glucuronosyltransferase 2B15 C-terminal domain L446S at 1.99 Angstroms resolution
To Be Published
|
|
1NCY
 
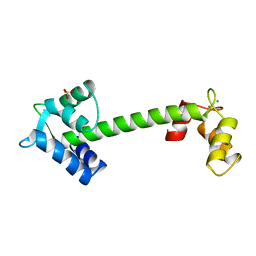 | | TROPONIN-C, COMPLEX WITH MANGANESE | | Descriptor: | MANGANESE (II) ION, SULFATE ION, TROPONIN C | | Authors: | Sundaralingam, M, Rao, S.T. | | Deposit date: | 1996-06-02 | | Release date: | 1997-01-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray structures of Mn, Cd and Tb metal complexes of troponin C.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1NCZ
 
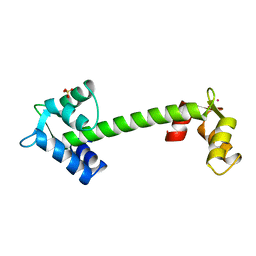 | | TROPONIN C | | Descriptor: | SULFATE ION, TERBIUM(III) ION, TROPONIN C | | Authors: | Sundaralingam, M, Rao, S.T. | | Deposit date: | 1996-06-02 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray structures of Mn, Cd and Tb metal complexes of troponin C.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1NCX
 
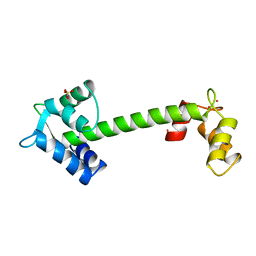 | | TROPONIN C | | Descriptor: | CADMIUM ION, SULFATE ION, TROPONIN C | | Authors: | Sundaralingam, M, Rao, S.T. | | Deposit date: | 1996-06-02 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray structures of Mn, Cd and Tb metal complexes of troponin C.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1NT2
 
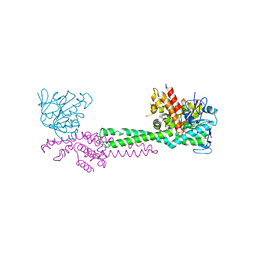 | | CRYSTAL STRUCTURE OF FIBRILLARIN/NOP5P COMPLEX | | Descriptor: | Fibrillarin-like pre-rRNA processing protein, S-ADENOSYLMETHIONINE, conserved hypothetical protein | | Authors: | Aittaleb, M, Rashid, R, Chen, Q, Palmer, J.R, Daniels, C.J, Li, H. | | Deposit date: | 2003-01-28 | | Release date: | 2003-04-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure and function of archaeal box C/D sRNP core proteins.
Nat.Struct.Biol., 10, 2003
|
|
3QR0
 
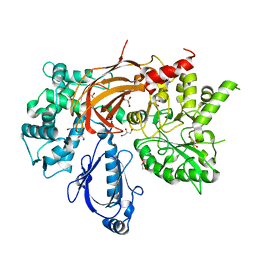 | | Crystal Structure of S. officinalis PLC21 | | Descriptor: | CALCIUM ION, GLYCEROL, phospholipase C-beta (PLC-beta) | | Authors: | Lyon, A.M, Northup, J.K, Tesmer, J.J.G. | | Deposit date: | 2011-02-16 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An autoinhibitory helix in the C-terminal region of phospholipase C-beta mediates Galphaq activation.
Nat.Struct.Mol.Biol., 18, 2011
|
|
4F09
 
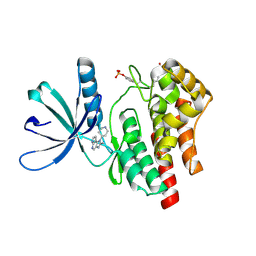 | |
1EWH
 
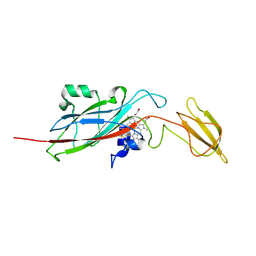 | | STRUCTURE OF CYTOCHROME F FROM CHLAMYDOMONAS REINHARDTII | | Descriptor: | ACETATE ION, CYTOCHROME F, HEME C | | Authors: | Sainz, G, Carrell, C.J, Ponamarev, M.V, Soriano, G.M, Cramer, W.A, Smith, J.L. | | Deposit date: | 2000-04-25 | | Release date: | 2000-08-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Interruption of the internal water chain of cytochrome f impairs photosynthetic function.
Biochemistry, 39, 2000
|
|
3IRW
 
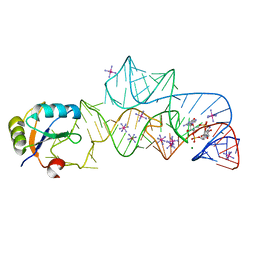 | | Structure of a c-di-GMP riboswitch from V. cholerae | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), IRIDIUM HEXAMMINE ION, MAGNESIUM ION, ... | | Authors: | Smith, K.D. | | Deposit date: | 2009-08-24 | | Release date: | 2009-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of ligand binding by a c-di-GMP riboswitch.
Nat.Struct.Mol.Biol., 16, 2009
|
|
1QN0
 
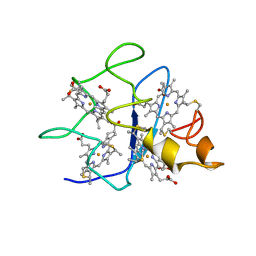 | | SOLUTION STRUCTURE OF DESULFOVIBRIO GIGAS FERROCYTOCHROME C3, NMR, 20 STRUCTURES | | Descriptor: | CYTOCHROME C3, HEME C | | Authors: | Messias, A.C, Teodoro, M.L, Brennan, L, Legall, J, Santos, H, Xavier, A.V, Turner, D.L. | | Deposit date: | 1999-10-11 | | Release date: | 2000-10-12 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for the Network of Functional Cooperativities in Cytochrome C3 from Desulfovibrio Gigas: Solution Structures of the Oxidised and Reduced States
J.Mol.Biol., 298, 2000
|
|
4NWK
 
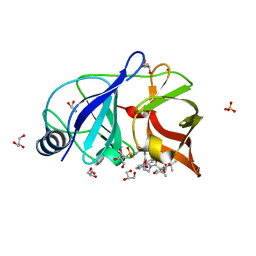 | | Crystal structure of hepatis c virus protease (ns3) complexed with bms-605339 aka n-(tert-butoxycarbonyl)-3-me thyl-l-valyl-(4r)-n-((1r,2s)-1-((cyclopropylsulfonyl)carba moyl)-2-vinylcyclopropyl)-4-((6-methoxy-1-isoquinolinyl)ox y)-l-prolinamide | | Descriptor: | GLYCEROL, HCV NS3 1a Protease, N-(tert-butoxycarbonyl)-3-methyl-L-valyl-(4R)-N-{(1R,2S)-1-[(cyclopropylsulfonyl)carbamoyl]-2-ethenylcyclopropyl}-4-[(6-methoxyisoquinolin-1-yl)oxy]-L-prolinamide, ... | | Authors: | Muckelbauer, J.K, Klei, H.E. | | Deposit date: | 2013-12-06 | | Release date: | 2014-03-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Discovery and Early Clinical Evaluation of BMS-605339, a Potent and Orally Efficacious Tripeptidic Acylsulfonamide NS3 Protease Inhibitor for the Treatment of Hepatitis C Virus Infection.
J.Med.Chem., 57, 2014
|
|
1Q9I
 
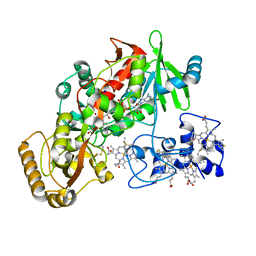 | | The A251C:S430C double mutant of flavocytochrome c3 from Shewanella frigidimarina | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, HEME C, MALATE LIKE INTERMEDIATE, ... | | Authors: | Rothery, E.L, Mowat, C.G, Miles, C.S, Walkinshaw, M.D, Reid, G.A, Chapman, S.K. | | Deposit date: | 2003-08-25 | | Release date: | 2004-05-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Probing Domain Mobility in a Flavocytochrome
Biochemistry, 42, 2004
|
|
1YRZ
 
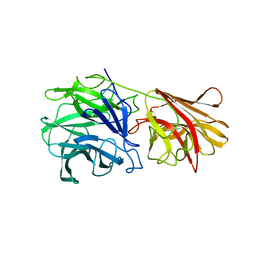 | | Crystal structure of xylan beta-1,4-xylosidase from Bacillus Halodurans C-125 | | Descriptor: | xylan beta-1,4-xylosidase | | Authors: | Fedorov, A.A, Fedorov, E.V, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-02-05 | | Release date: | 2005-02-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of xylan beta-1,4-xylosidase from Bacillus Halodurans C-125
To be Published
|
|
4DT7
 
 | | Crystal structure of thrombin bound to the activation domain QEDQVDPRLIDGKMTRRGDS of protein C | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, SODIUM ION, ... | | Authors: | Pozzi, N, Barranco-Medina, S, Chen, Z, Di Cera, E. | | Deposit date: | 2012-02-20 | | Release date: | 2012-05-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Exposure of R169 controls protein C activation and autoactivation.
Blood, 120, 2012
|
|
1CPG
 
 | |
1CPE
 
 | |
1LC2
 
 | |
