1J2B
 
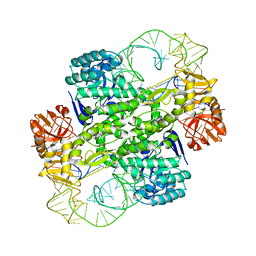 | | Crystal Structure Of Archaeosine tRNA-Guanine Transglycosylase Complexed With lambda-form tRNA(Val) | | Descriptor: | Archaeosine tRNA-guanine transglycosylase, MAGNESIUM ION, ZINC ION, ... | | Authors: | Ishitani, R, Nureki, O, Nameki, N, Okada, N, Nishimura, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-12-29 | | Release date: | 2003-05-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Alternative Tertiary Structure of tRNA for Recognition by a Posttranscriptional Modification Enzyme
Cell(Cambridge,Mass.), 113, 2003
|
|
5DO4
 
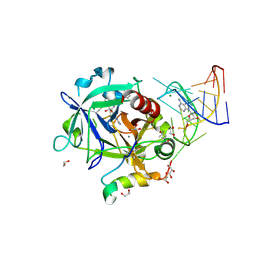 | | Thrombin-RNA aptamer complex | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2015-09-10 | | Release date: | 2016-09-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.859 Å) | | Cite: | Evoking picomolar binding in RNA by a single phosphorodithioate linkage.
Nucleic Acids Res., 44, 2016
|
|
3CDJ
 
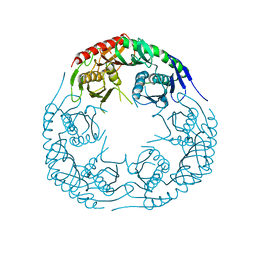 | | Crystal structure of the E. coli KH/S1 domain truncated PNPase | | Descriptor: | Polynucleotide phosphorylase | | Authors: | Shi, Z, Yang, W.Z, Lin-Chao, S, Chak, K.F, Yuan, H.S. | | Deposit date: | 2008-02-27 | | Release date: | 2008-12-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of Escherichia coli PNPase: central channel residues are involved in processive RNA degradation.
Rna, 14, 2008
|
|
9FVD
 
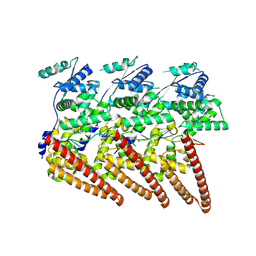 | | Cryo-EM structure of single-layered nucleoprotein-RNA helical assembly from Marburg virus, trimeric repeat unit | | Descriptor: | Nucleoprotein, RNA (5'-R(P*AP*GP*AP*CP*AP*CP*AP*CP*AP*AP*AP*AP*AP*CP*AP*AP*GP*A)-3') | | Authors: | Zinzula, L, Beck, F, Camasta, M, Bohn, S, Liu, C, Morado, D, Bracher, A, Plitzko, J.M, Baumeister, W. | | Deposit date: | 2024-06-26 | | Release date: | 2024-10-09 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of single-layered nucleoprotein-RNA complex from Marburg virus.
Nat Commun, 15, 2024
|
|
2HGZ
 
 | | Crystal structure of a p-benzoyl-L-phenylalanyl-tRNA synthetase | | Descriptor: | PARA-(BENZOYL)-PHENYLALANINE, Tyrosyl-tRNA synthetase | | Authors: | Liu, W, Alfonta, L, Mack, A.V, Schultz, P.G. | | Deposit date: | 2006-06-27 | | Release date: | 2007-06-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the recognition of para-benzoyl-L-phenylalanine by evolved aminoacyl-tRNA synthetases.
Angew.Chem.Int.Ed.Engl., 46, 2007
|
|
5J0M
 
 | | Ground state sampled during RDC restrained Replica-averaged Metadynamics (RAM) simulations of the HIV-1 TAR complexed with cyclic peptide mimetic of Tat | | Descriptor: | Apical region (29-mer) of the HIV-1 TAR RNA element, Cyclic peptide mimetic of HIV-1 Tat | | Authors: | Borkar, A.N, Bardaro Jr, M.F, Varani, G, Vendruscolo, M. | | Deposit date: | 2016-03-28 | | Release date: | 2016-06-08 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Structure of a low-population binding intermediate in protein-RNA recognition.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
2LBR
 
 | |
1AM0
 
 | | AMP RNA APTAMER COMPLEX, NMR, 8 STRUCTURES | | Descriptor: | ADENOSINE MONOPHOSPHATE, RNA APTAMER | | Authors: | Jiang, F, Kumar, R.A, Jones, R.A, Patel, D.J. | | Deposit date: | 1997-06-19 | | Release date: | 1997-07-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of RNA Folding and Recognition in an AMP-RNA Aptamer Complex
Nature, 382, 1996
|
|
7B2I
 
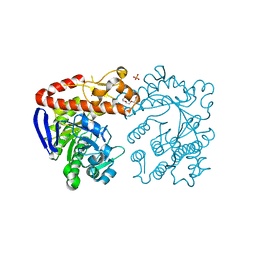 | | Heterodimeric tRNA-Guanine Transglycosylase from mouse | | Descriptor: | CITRIC ACID, DI(HYDROXYETHYL)ETHER, HEXAETHYLENE GLYCOL, ... | | Authors: | Sebastiani, M, Heine, A, Reuter, K. | | Deposit date: | 2020-11-27 | | Release date: | 2021-12-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and Biochemical Investigation of the Heterodimeric Murine tRNA-Guanine Transglycosylase.
Acs Chem.Biol., 17, 2022
|
|
5J2W
 
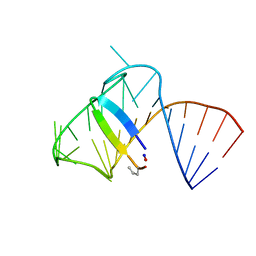 | | Intermediate state lying on the pathway of release of Tat from HIV-1 TAR. | | Descriptor: | Apical region (29mer) of the HIV-1 TAR RNA element, Cyclic peptide mimetic of HIV-1 Tat | | Authors: | Borkar, A.N, Bardaro Jr, M.F, Varani, G, Vendruscolo, M. | | Deposit date: | 2016-03-30 | | Release date: | 2016-06-08 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Structure of a low-population binding intermediate in protein-RNA recognition.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1FQZ
 
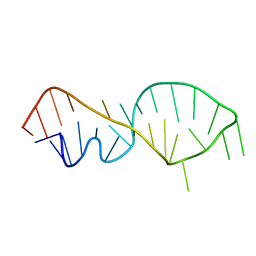 | | NMR VALIDATED MODEL OF DOMAIN IIID OF HEPATITIS C VIRUS INTERNAL RIBOSOME ENTRY SITE | | Descriptor: | HEPATITIS C VIRUS IRES DOMAIN IIID | | Authors: | Klinck, R, Westhof, E, Walker, S, Afshar, M, Collier, A, Aboul-ela, F. | | Deposit date: | 2000-09-07 | | Release date: | 2001-01-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A potential RNA drug target in the hepatitis C virus internal ribosomal entry site.
RNA, 6, 2000
|
|
6Z9U
 
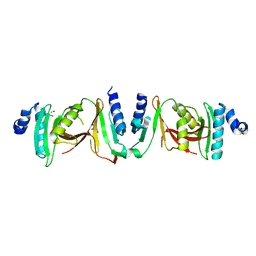 | | Crystal structure of a TSEN15-34 heterodimer. | | Descriptor: | GLYCEROL, tRNA-splicing endonuclease subunit Sen15, tRNA-splicing endonuclease subunit Sen34 | | Authors: | Trowitzsch, S, Sekulovski, S. | | Deposit date: | 2020-06-04 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.10001779 Å) | | Cite: | Assembly defects of human tRNA splicing endonuclease contribute to impaired pre-tRNA processing in pontocerebellar hypoplasia.
Nat Commun, 12, 2021
|
|
1JU7
 
 | |
1JWC
 
 | |
1GTN
 
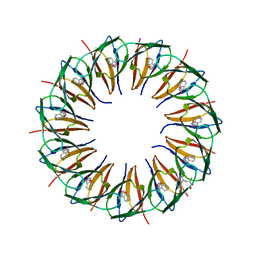 | | Structure of the trp RNA-binding attenuation protein (TRAP) bound to an RNA molecule containing 11 GAGCC repeats | | Descriptor: | (GAGCC)11G 56-NUCLEOTIDE RNA, TRP RNA-BINDING ATTENUATION PROTEIN, TRYPTOPHAN | | Authors: | Hopcroft, N.H, Wendt, A.L, Gollnick, P, Antson, A.A. | | Deposit date: | 2002-01-16 | | Release date: | 2002-04-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Specificity of Trap-RNA Interactions: Crystal Structures of Two Complexes with Different RNA Sequences
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1GTF
 
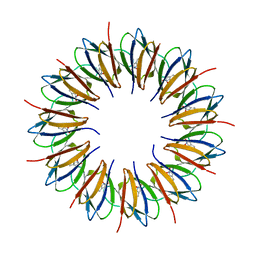 | | The structure of the trp RNA-binding attenuation protein (TRAP) bound to a 53-nucleotide RNA molecule containing GAGUU repeats | | Descriptor: | (GAGUU)10GAG 53-NUCLEOTIDE RNA, TRP RNA-BINDING ATTENUATION PROTEIN (TRAP), TRYPTOPHAN | | Authors: | Hopcroft, N.H, Wendt, A.L, Gollnick, P, Antson, A.A. | | Deposit date: | 2002-01-15 | | Release date: | 2002-04-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Specificity of Trap-RNA Interactions: Crystal Structures of Two Complexes with Different RNA Sequences
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1Y2Y
 
 | | Structural Characterization of Nop10p using Nuclear Magnetic Resonance Spectroscopy | | Descriptor: | Ribosome biogenesis protein Nop10 | | Authors: | Khanna, M, Wu, H, Johansson, C, Caizergues-Ferrer, M, Feigon, J. | | Deposit date: | 2004-11-23 | | Release date: | 2005-12-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural study of the H/ACA snoRNP components Nop10p and the 3' hairpin of U65 snoRNA
RNA, 12, 2006
|
|
7QDD
 
 | |
7QDE
 
 | |
3RG5
 
 | | Crystal Structure of Mouse tRNA(Sec) | | Descriptor: | ACETATE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Wahl, M.C, Ganichkin, O.M, Anedchenko, E.A. | | Deposit date: | 2011-04-07 | | Release date: | 2011-06-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure analysis reveals functional flexibility in the selenocysteine-specific tRNA from mouse.
Plos One, 6, 2011
|
|
1HC8
 
 | | CRYSTAL STRUCTURE OF A CONSERVED RIBOSOMAL PROTEIN-RNA COMPLEX | | Descriptor: | 58 NUCLEOTIDE RIBOSOMAL 23S RNA DOMAIN, MAGNESIUM ION, OSMIUM ION, ... | | Authors: | Conn, G.L, Draper, D.E, Lattman, E.E, Gittis, A.G. | | Deposit date: | 2001-04-27 | | Release date: | 2002-05-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A Compact RNA Tertiary Structure Contains a Buried Backbone-K+ Complex
J.Mol.Biol., 318, 2002
|
|
1ETG
 
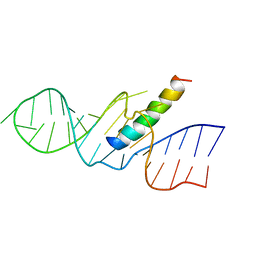 | | REV RESPONSE ELEMENT (RRE) RNA COMPLEXED WITH REV PEPTIDE, NMR, 19 STRUCTURES | | Descriptor: | REV PEPTIDE, REV RESPONSIVE ELEMENT RNA | | Authors: | Battiste, J.L, Mao, H, Rao, N.S, Tan, R, Muhandiram, D.R, Kay, L.E, Frankel, A.D, Willamson, J.R. | | Deposit date: | 1996-08-28 | | Release date: | 1997-03-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Alpha helix-RNA major groove recognition in an HIV-1 rev peptide-RRE RNA complex.
Science, 273, 1996
|
|
1ETF
 
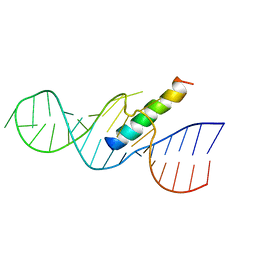 | | REV RESPONSE ELEMENT (RRE) RNA COMPLEXED WITH REV PEPTIDE, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | REV PEPTIDE, REV RESPONSE ELEMENT RNA | | Authors: | Battiste, J.L, Mao, H, Rao, N.S, Tan, R, Muhandiram, D.R, Kay, L.E, Frankel, A.D, Willamson, J.R. | | Deposit date: | 1996-08-28 | | Release date: | 1997-03-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Alpha helix-RNA major groove recognition in an HIV-1 rev peptide-RRE RNA complex.
Science, 273, 1996
|
|
4NL2
 
 | |
2XDD
 
 | | A processed non-coding RNA regulates a bacterial antiviral system | | Descriptor: | TOXI, TOXN, ZINC ION | | Authors: | Blower, T.R, Pei, X.Y, Short, F.L, Fineran, P.C, Humphreys, D.P, Luisi, B.F, Salmond, G.P.C. | | Deposit date: | 2010-04-30 | | Release date: | 2011-01-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The Phage Abortive Infection System, Toxin, Functions as a Protein-RNA Toxin-Antitoxin Pair.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
