6HJK
 
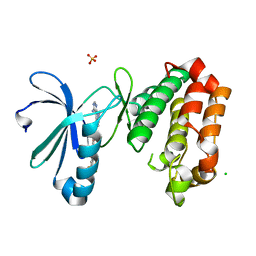 | | Crystal Structure of Aurora-A L210C catalytic domain in complex with ASDO2 | | Descriptor: | (~{E})-~{N}-[4-(4-azanyl-1-propan-2-yl-pyrazolo[3,4-d]pyrimidin-3-yl)phenyl]-4-[4-fluoranyl-3-(trifluoromethyl)phenyl]-4-oxidanylidene-but-2-enamide, Aurora kinase A, CHLORIDE ION, ... | | Authors: | Bayliss, R, McIntyre, P.J. | | Deposit date: | 2018-09-04 | | Release date: | 2018-10-03 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Type II Kinase Inhibitors Targeting Cys-Gatekeeper Kinases Display Orthogonality with Wild Type and Ala/Gly-Gatekeeper Kinases.
ACS Chem. Biol., 13, 2018
|
|
5RA4
 
 | |
9ECO
 
 | |
5MMN
 
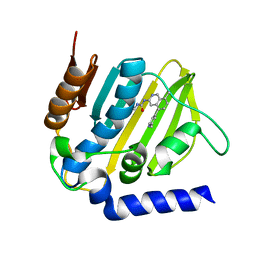 | | E. coli DNA Gyrase B 24 kDa ATPase domain in complex with 1-ethyl-3-[8-methyl-5-(2-methyl-pyridin-4-yl)-isoquinolin-3-yl]-urea | | Descriptor: | 1-ethyl-3-[8-methyl-5-(2-methylpyridin-4-yl)isoquinolin-3-yl]urea, DNA gyrase subunit B | | Authors: | Panchaud, P, Bruyere, T, Blumstein, A.-C, Bur, D, Chambovey, A, Ertel, E.A, Gude, M, Hubschwerlen, C, Jacob, L, Kimmerlin, T, Pfeifer, T, Prade, L, Seiler, P, Ritz, D, Rueedi, G. | | Deposit date: | 2016-12-12 | | Release date: | 2017-04-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery and Optimization of Isoquinoline Ethyl Ureas as Antibacterial Agents.
J. Med. Chem., 60, 2017
|
|
8SUB
 
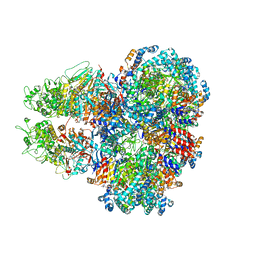 | | E. coli SIR2-HerA complex (dodecamer SIR2 pentamer HerA) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Nucleoside triphosphate hydrolase, ... | | Authors: | Shen, Z.F, Lin, Q.P, Fu, T.M. | | Deposit date: | 2023-05-11 | | Release date: | 2023-12-27 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Assembly-mediated activation of the SIR2-HerA supramolecular complex for anti-phage defense.
Mol.Cell, 83, 2023
|
|
4PNC
 
 | | E. COLI METHIONINE AMINOPEPTIDASE IN COMPLEX WITH INHIBITOR 7-METHOXY-2-METHYLEN-3,4-DIHYDRONAPHTHALEN-1(2H)-ONE | | Descriptor: | (2S)-7-methoxy-2-methyl-3,4-dihydronaphthalen-1(2H)-one, COBALT (II) ION, Methionine aminopeptidase, ... | | Authors: | Scheidig, A.J, Altmeyer, M, Klein, C.D. | | Deposit date: | 2014-05-23 | | Release date: | 2014-07-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Beta-aminoketones as prodrugs for selective irreversible inhibitors of type-1 methionine aminopeptidases.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
8UAE
 
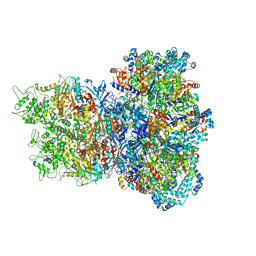 | | E. coli Sir2_HerA complex (12:6) with ATPgamaS | | Descriptor: | MAGNESIUM ION, Nucleoside triphosphate hydrolase, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Shen, Z.F, Lin, Q.P, Fu, T.M. | | Deposit date: | 2023-09-20 | | Release date: | 2023-12-27 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Assembly-mediated activation of the SIR2-HerA supramolecular complex for anti-phage defense.
Mol.Cell, 83, 2023
|
|
8TXO
 
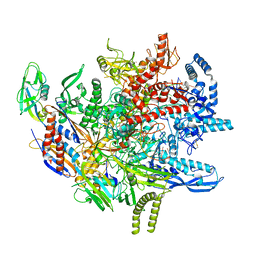 | | E. coli DNA-directed RNA polymerase transcription elongation complex bound to the unnatural dZ-PTP base pair in the active site | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Aiyer, S, Shan, Z, Oh, J, Wang, D, Tan, Y.Z, Lyumkis, D. | | Deposit date: | 2023-08-23 | | Release date: | 2024-01-24 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Overcoming resolution attenuation during tilted cryo-EM data collection.
Nat Commun, 15, 2024
|
|
1MMI
 
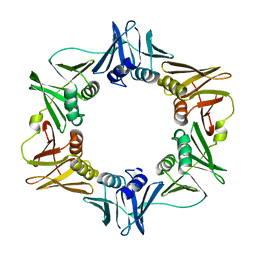 | | E. COLI DNA POLYMERASE BETA SUBUNIT | | Descriptor: | DNA polymerase III, beta chain | | Authors: | Oakley, A.J, Prosselkov, P, Wijffels, G, Beck, J.L, Wilce, M.C.J, Dixon, N.E. | | Deposit date: | 2002-09-04 | | Release date: | 2003-09-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.848 Å) | | Cite: | Flexibility revealed by the 1.85 A crystal structure of the beta sliding-clamp subunit of Escherichia coli DNA polymerase III.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1KH9
 
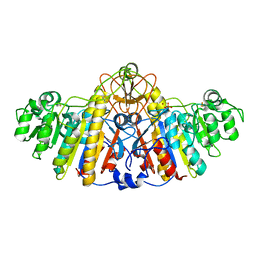 | | E. COLI ALKALINE PHOSPHATASE MUTANT (D153GD330N) COMPLEX WITH PHOSPHATE | | Descriptor: | Alkaline phosphatase, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Le Du, M.H, Lamoure, C, Muller, B.H, Bulgakov, O.V, Lajeunesse, E. | | Deposit date: | 2001-11-29 | | Release date: | 2002-03-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Artificial evolution of an enzyme active site: structural studies of three highly active mutants of Escherichia coli alkaline phosphatase.
J.Mol.Biol., 316, 2002
|
|
1KHK
 
 | | E. COLI ALKALINE PHOSPHATASE MUTANT (D153HD330N) | | Descriptor: | Alkaline Phosphatase, MAGNESIUM ION, ZINC ION | | Authors: | Le Du, M.H, Lamoure, C, Muller, B.H, Bulgakov, O.V, Lajeunesse, E, Menez, A, Boulain, J.C. | | Deposit date: | 2001-11-30 | | Release date: | 2002-03-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Artificial evolution of an enzyme active site: structural studies of three highly active mutants of Escherichia coli alkaline phosphatase.
J.Mol.Biol., 316, 2002
|
|
1KH7
 
 | | E. COLI ALKALINE PHOSPHATASE MUTANT (D153GD330N) | | Descriptor: | MAGNESIUM ION, SULFATE ION, ZINC ION, ... | | Authors: | Le Du, M.H, Lamoure, C, Muller, B.H, Bulgakov, O.V, Lajeunesse, E. | | Deposit date: | 2001-11-29 | | Release date: | 2002-03-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Artificial evolution of an enzyme active site: structural studies of three highly active mutants of Escherichia coli alkaline phosphatase.
J.Mol.Biol., 316, 2002
|
|
1KHL
 
 | | E. COLI ALKALINE PHOSPHATASE MUTANT (D153HD330N) COMPLEX WITH PHOSPHATE | | Descriptor: | Alkaline Phosphatase, PHOSPHATE ION, ZINC ION | | Authors: | Le Du, M.H, Lamoure, C, Muller, B.H, Bulgakov, O.V, Lajeunesse, E, Menez, A, Boulain, J.C. | | Deposit date: | 2001-11-30 | | Release date: | 2002-03-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Artificial evolution of an enzyme active site: structural studies of three highly active mutants of Escherichia coli alkaline phosphatase.
J.Mol.Biol., 316, 2002
|
|
1PE6
 
 | | REFINED X-RAY STRUCTURE OF PAPAIN(DOT)E-64-C COMPLEX AT 2.1-ANGSTROMS RESOLUTION | | Descriptor: | METHANOL, N-[1-HYDROXYCARBOXYETHYL-CARBONYL]LEUCYLAMINO-2-METHYL-BUTANE, PAPAIN | | Authors: | Yamamoto, D, Matsumoto, K, Ohishi, H, Ishida, T, Inoue, M, Kitamura, K, Mizuno, H. | | Deposit date: | 1991-05-14 | | Release date: | 1993-04-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Refined x-ray structure of papain.E-64-c complex at 2.1-A resolution.
J.Biol.Chem., 266, 1991
|
|
1KHN
 
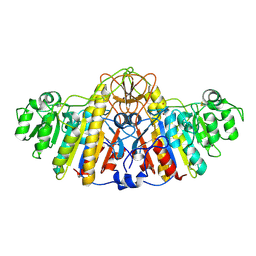 | | E. COLI ALKALINE PHOSPHATASE MUTANT (D153HD330N) ZINC FORM | | Descriptor: | Alkaline phosphatase, ZINC ION | | Authors: | Le Du, M.H, Lamoure, C, Muller, B.H, Bulgakov, O.V, Lajeunesse, E, Menez, A, Boulain, J.C. | | Deposit date: | 2001-11-30 | | Release date: | 2002-03-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Artificial evolution of an enzyme active site: structural studies of three highly active mutants of Escherichia coli alkaline phosphatase.
J.Mol.Biol., 316, 2002
|
|
2PIC
 
 | |
1KH4
 
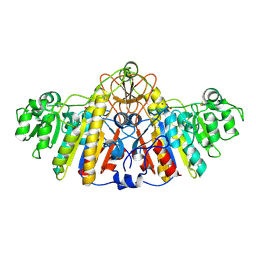 | | E. COLI ALKALINE PHOSPHATASE MUTANT (D330N) IN COMPLEX WITH PHOSPHATE | | Descriptor: | ALKALINE PHOSPHATASE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Le Du, M.H, Lamoure, C, Muller, B.H, Bulgakov, O.V, Lajeunesse, E. | | Deposit date: | 2001-11-29 | | Release date: | 2002-03-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Artificial evolution of an enzyme active site: structural studies of three highly active mutants of Escherichia coli alkaline phosphatase.
J.Mol.Biol., 316, 2002
|
|
1LN4
 
 | |
5U6R
 
 | |
1QFG
 
 | | E. COLI FERRIC HYDROXAMATE RECEPTOR (FHUA) | | Descriptor: | 3-HYDROXY-TETRADECANOIC ACID, DECYLAMINE-N,N-DIMETHYL-N-OXIDE, DIPHOSPHATE, ... | | Authors: | Ferguson, A.D, Welte, W, Hofmann, E, Lindner, B, Holst, O, Coulton, J.W, Diederichs, K. | | Deposit date: | 1999-04-10 | | Release date: | 2000-07-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A conserved structural motif for lipopolysaccharide recognition by procaryotic and eucaryotic proteins.
Structure Fold.Des., 8, 2000
|
|
4E7G
 
 | |
4E7F
 
 | | E. cloacae C115D MurA in complex with UDP | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, UDP-N-acetylglucosamine 1-carboxyvinyltransferase, ... | | Authors: | Zhu, J.-Y, Betzi, S, Yang, Y, Schonbrunn, E. | | Deposit date: | 2012-03-16 | | Release date: | 2013-03-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Open-close transition of MurA
To be Published
|
|
4Q65
 
 | | Structure of the E. coli Peptide Transporter YbgH | | Descriptor: | Dipeptide permease D | | Authors: | Zhang, C, Zhao, Y, Mao, G, Liu, M, Wang, X. | | Deposit date: | 2014-04-21 | | Release date: | 2014-08-13 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal structure of the E. coli peptide transporter YbgH.
Structure, 22, 2014
|
|
1QFF
 
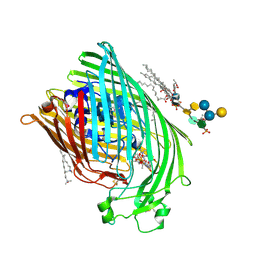 | | E. COLI FERRIC HYDROXAMATE UPTAKE RECEPTOR (FHUA) IN COMPLEX WITH BOUND FERRICHROME-IRON | | Descriptor: | 2-AMINO-VINYL-PHOSPHATE, 3-HYDROXY-TETRADECANOIC ACID, DECYLAMINE-N,N-DIMETHYL-N-OXIDE, ... | | Authors: | Ferguson, A.D, Hofmann, E, Coulton, J.W, Diederichs, K, Welte, W. | | Deposit date: | 1999-04-10 | | Release date: | 2000-07-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A conserved structural motif for lipopolysaccharide recognition by procaryotic and eucaryotic proteins.
Structure Fold.Des., 8, 2000
|
|
2PQ9
 
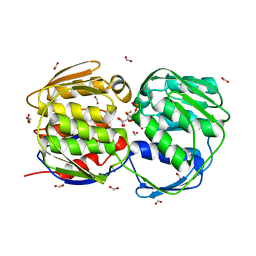 | | E. coli EPSPS liganded with (R)-difluoromethyl tetrahedral reaction intermediate analog | | Descriptor: | (3R,4S,5R)-5-[(1R)-1-CARBOXY-2,2-DIFLUORO-1-(PHOSPHONOOXY)ETHOXY]-4-HYDROXY-3-(PHOSPHONOOXY)CYCLOHEX-1-ENE-1-CARBOXYLIC ACID, 3-phosphoshikimate 1-carboxyvinyltransferase, FORMIC ACID | | Authors: | Healy-Fried, M.L, Funke, T, Han, H, Schonbrunn, E. | | Deposit date: | 2007-05-01 | | Release date: | 2008-03-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Differential inhibition of class I and class II 5-enolpyruvylshikimate-3-phosphate synthases by tetrahedral reaction intermediate analogues.
Biochemistry, 46, 2007
|
|
