1ESH
 
 | |
2AWX
 
 | | Synapse associated protein 97 PDZ2 domain variant C378S | | Descriptor: | HISTIDINE, Synapse associated protein 97 | | Authors: | Von Ossowski, I, Oksanen, E, Von Ossowski, L, Cai, C, Sundberg, M, Goldman, A, Keinanen, K. | | Deposit date: | 2005-09-02 | | Release date: | 2006-08-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the second PDZ domain of SAP97 in complex with a GluR-A C-terminal peptide
Febs J., 273, 2006
|
|
2LIZ
 
 | |
2JJR
 
 | | V232K, N236D-trichosanthin | | Descriptor: | DI(HYDROXYETHYL)ETHER, RIBOSOME-INACTIVATING PROTEIN ALPHA-TRICHOSANTHIN, SULFATE ION, ... | | Authors: | Too, P.H, Ma, M.K, Mak, A.N, Tung, C.K, Zhu, G, Au, S.W, Wong, K.B, Shaw, P.C. | | Deposit date: | 2008-04-21 | | Release date: | 2008-12-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The C-Terminal Fragment of the Ribosomal P Protein Complexed to Trichosanthin Reveals the Interaction between the Ribosome-Inactivating Protein and the Ribosome.
Nucleic Acids Res., 37, 2009
|
|
2IWQ
 
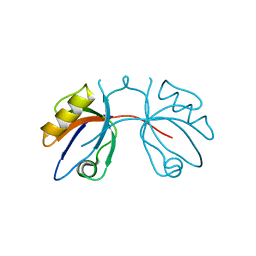 | | 7th PDZ domain of Multiple PDZ Domain Protein MPDZ | | Descriptor: | MULTIPLE PDZ DOMAIN PROTEIN | | Authors: | Elkins, J.M, Berridge, G, Savitsky, P, Smee, C.E.A, Turnbull, A, Ugochukwu, E, Papagrigoriou, E, Sundstrom, M, Edwards, A, Arrowsmith, C, Weigelt, J, Doyle, D.A. | | Deposit date: | 2006-07-03 | | Release date: | 2006-07-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Pick1 and Other Pdz Domains Obtained with the Help of Self-Binding C-Terminal Extensions.
Protein Sci., 16, 2007
|
|
4DN0
 
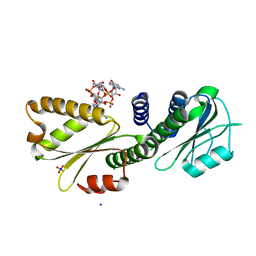 | | PelD 156-455 from Pseudomonas aeruginosa PA14 in complex with c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Putative uncharacterized protein pelD, SODIUM ION, ... | | Authors: | Whitney, J.C, Colvin, K.M, Marmont, L.S, Robinson, H, Parsek, M.R, Howell, P.L. | | Deposit date: | 2012-02-08 | | Release date: | 2012-05-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the Cytoplasmic Region of PelD, a Degenerate Diguanylate Cyclase Receptor That Regulates Exopolysaccharide Production in Pseudomonas aeruginosa.
J.Biol.Chem., 287, 2012
|
|
4AE2
 
 | | Crystal structure of Human fibrillar procollagen type III C- propeptide trimer | | Descriptor: | CALCIUM ION, COLLAGEN ALPHA-1(III) CHAIN, NITRATE ION | | Authors: | Bourhis, J.M, Mariano, N, Zhao, Y, Harlos, K, Jones, E.Y, Moali, C, Aghajari, N, Hulmes, D.J. | | Deposit date: | 2012-01-05 | | Release date: | 2012-09-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural Basis of Fibrillar Collagen Trimerization and Related Genetic Disorders.
Nat.Struct.Mol.Biol., 19, 2012
|
|
1H21
 
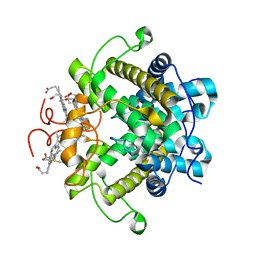 | | A novel iron centre in the split-Soret cytochrome c from Desulfovibrio desulfuricans ATCC 27774 | | Descriptor: | HEME C, SPLIT-SORET CYTOCHROME C | | Authors: | Abreu, I.A, Lourenco, A.I, Xavier, A.V, Legall, J, Coelho, A.V, Matias, P.M, Pinto, D.M, Carrondo, M.A, Teixeira, M, Saraiva, L.M. | | Deposit date: | 2002-07-30 | | Release date: | 2003-02-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Novel Iron Centre in the Split-Soret Cytochrome C from Desulfovibrio Desulfuricans Atcc 27774
J.Biol.Inorg.Chem., 8, 2003
|
|
2BH5
 
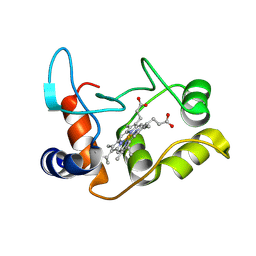 | | X-ray structure of the M100K variant of ferric cyt c-550 from Paracoccus versutus determined at 295 K. | | Descriptor: | CYTOCHROME C-550, HEME C | | Authors: | Worrall, J.A.R, van Roon, A.-M.M, Ubbink, M, Canters, G.W. | | Deposit date: | 2005-01-07 | | Release date: | 2005-05-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Effect of Replacing the Axial Methionine Ligand with a Lysine Residue in Cytochrome C-550 from Paracoccus Versutus Assessed by X-Ray Crystallography and Unfolding.
FEBS J., 272, 2005
|
|
4P5H
 
 | |
2O1L
 
 | | Structure of a complex of C-terminal lobe of bovine lactoferrin with disaccharide at 1.97 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CARBONATE ION, FE (III) ION, ... | | Authors: | Singh, N, Sharma, S, Perbandt, M, Kaur, P, Betzel, C, Singh, T.P. | | Deposit date: | 2006-11-29 | | Release date: | 2006-12-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structure of a complex of C-terminal lobe of bovine lactoferrin with disaccharide at 1.97 A resolution
To be Published
|
|
2JUJ
 
 | |
3B76
 
 | | Crystal structure of the third PDZ domain of human ligand-of-numb protein-X (LNX1) in complex with the C-terminal peptide from the coxsackievirus and adenovirus receptor | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase LNX, SODIUM ION | | Authors: | Ugochukwu, E, Burgess-Brown, N, Berridge, G, Elkins, J, Bunkoczi, G, Pike, A.C.W, Sundstrom, M, Arrowsmith, C.H, Weigelt, J, Edwards, A.M, Gileadi, O, von Delft, F, Doyle, D, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-10-30 | | Release date: | 2007-11-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the third PDZ domain of human ligand-of-numb protein-X (LNX1) in complex with the C-terminal peptide from the coxsackievirus and adenovirus receptor.
To be Published
|
|
4AIF
 
 | | AIP TPR domain in complex with human Hsp90 peptide | | Descriptor: | AH RECEPTOR-INTERACTING PROTEIN, HEAT SHOCK PROTEIN HSP 90-ALPHA, SULFATE ION | | Authors: | Morgan, R.M.L, Roe, S.M, Pearl, L.H, Prodromou, C. | | Deposit date: | 2012-02-09 | | Release date: | 2013-01-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.006 Å) | | Cite: | Structure of the Tpr Domain of Aip: Lack of Client Protein Interaction with the C-Terminal Alpha-7 Helix of the Tpr Domain of Aip is Sufficient for Pituitary Adenoma Predisposition.
Plos One, 7, 2012
|
|
3IHO
 
 | | The C-terminal glycosylase domain of human MBD4 | | Descriptor: | Methyl-CpG-binding domain protein 4 | | Authors: | Amaya, M.F, Xu, C, Bian, C.B, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-07-30 | | Release date: | 2009-08-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The C-terminal glycosylase domain of human MBD4
To be Published
|
|
3G2V
 
 | | VHS Domain of human GGA1 complexed with Sotilin C-terminal Phosphopeptide | | Descriptor: | ADP-ribosylation factor-binding protein GGA1, C-terminal fragment of Sortilin, IODIDE ION | | Authors: | Cramer, J.F, Behrens, M.A, Gustafsen, C, Oliveira, C.L.P, Pedersen, J.S, Madsen, P, Petersen, C.M, Thirup, S.S. | | Deposit date: | 2009-02-01 | | Release date: | 2009-12-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | GGA autoinhibition revisited
Traffic, 11, 2010
|
|
2M13
 
 | | The ZZ domain of cytoplasmic polyadenylation element binding protein 1 (CPEB1) | | Descriptor: | Cytoplasmic polyadenylation element-binding protein 1, ZINC ION | | Authors: | Lee, B.M, Merkel, D.J, Wells, S.B, Hilburn, B.C, Elazzouzi, F, Perez-Alvarado, G.C. | | Deposit date: | 2012-11-14 | | Release date: | 2013-04-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The C-Terminal Region of Cytoplasmic Polyadenylation Element Binding Protein Is a ZZ Domain with Potential for Protein-Protein Interactions.
J.Mol.Biol., 425, 2013
|
|
4QDR
 
 | |
1CNO
 
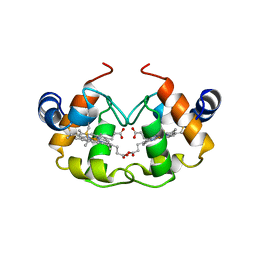 | | STRUCTURE OF PSEUDOMONAS NAUTICA CYTOCHROME C552, BY MAD METHOD | | Descriptor: | CYTOCHROME C552, GLYCEROL, HEME C | | Authors: | Brown, K, Nurizzo, D, Cambillau, C. | | Deposit date: | 1998-08-03 | | Release date: | 1999-07-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | MAD structure of Pseudomonas nautica dimeric cytochrome c552 mimicks the c4 Dihemic cytochrome domain association.
J.Mol.Biol., 289, 1999
|
|
1DW0
 
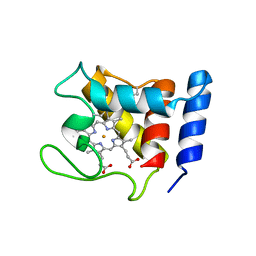 | | STRUCTURE OF OXIDIZED SHP, AN OXYGEN BINDING CYTOCHROME C | | Descriptor: | CYTOCHROME C, HEME C, SULFATE ION | | Authors: | Leys, D, Backers, K, Meyer, T.E, Hagen, W.R, Cusanovich, M.A, Van Beeumen, J.J. | | Deposit date: | 2000-01-24 | | Release date: | 2000-06-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structures of an oxygen-binding cytochrome c from Rhodobacter sphaeroides.
J.Biol.Chem., 275, 2000
|
|
1DW3
 
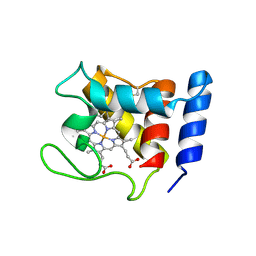 | | STRUCTURE OF A REDUCED OXYGEN BINDING CYTOCHROME C | | Descriptor: | CYTOCHROME C, HEME C | | Authors: | Leys, D, Backers, K, Meyer, T.E, Hagen, W.R, Cusanovich, M.A, Van Beeumen, J.J. | | Deposit date: | 2000-01-24 | | Release date: | 2000-06-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of an oxygen-binding cytochrome c from Rhodobacter sphaeroides.
J.Biol.Chem., 275, 2000
|
|
3O5A
 
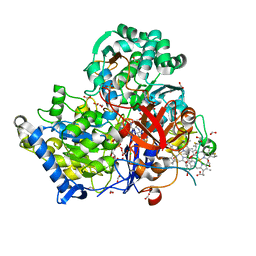 | | Crystal Structure of partially reduced Periplasmic Nitrate Reductase from Cupriavidus necator using Ionic Liquids | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, CHLORIDE ION, DIOXOTHIOMOLYBDENUM(VI) ION, ... | | Authors: | Coelho, C, Trincao, J, Romao, M.J. | | Deposit date: | 2010-07-28 | | Release date: | 2011-04-06 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | The crystal structure of Cupriavidus necator nitrate reductase in oxidized and partially reduced states
J.Mol.Biol., 408, 2011
|
|
1W25
 
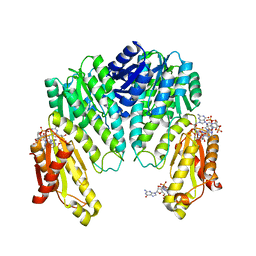 | | Response regulator PleD in complex with c-diGMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), MAGNESIUM ION, STALKED-CELL DIFFERENTIATION CONTROLLING PROTEIN, ... | | Authors: | Chan, C, Schirmer, T, Jenal, U. | | Deposit date: | 2004-06-28 | | Release date: | 2004-11-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis of Activity and Allosteric Control of Diguanylate Cyclase
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
7EBL
 
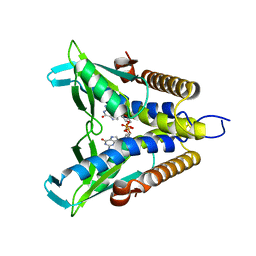 | | Bacterial STING in complex with c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), STING | | Authors: | Ko, T.-P, Wang, Y.-C, Yang, C.-S, Hou, M.-H, Chen, Y. | | Deposit date: | 2021-03-10 | | Release date: | 2021-09-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Crystal structure and functional implication of bacterial STING.
Nat Commun, 13, 2022
|
|
7EBD
 
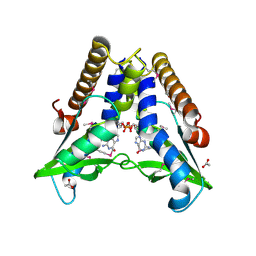 | | Bacterial STING in complex with c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), ACETATE ION, TIR-like domain-containing protein | | Authors: | Ko, T.-P, Wang, Y.-C, Yang, C.-S, Hou, M.-H, Chen, Y. | | Deposit date: | 2021-03-09 | | Release date: | 2021-09-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure and functional implication of bacterial STING.
Nat Commun, 13, 2022
|
|
