5L01
 
 | | Tryptophan 5-hydroxylase in complex with inhibitor (3~{S})-8-[2-azanyl-6-[(1~{R})-1-(4-chloranyl-2-phenyl-phenyl)-2,2,2-tris(fluoranyl)ethoxy]pyrimidin-4-yl]-2,8-diazaspiro[4.5]decane-3-carboxylic acid | | Descriptor: | (3~{S})-8-[2-azanyl-6-[(1~{R})-1-(4-chloranyl-2-phenyl-phenyl)-2,2,2-tris(fluoranyl)ethoxy]pyrimidin-4-yl]-2,8-diazaspiro[4.5]decane-3-carboxylic acid, FE (III) ION, Tryptophan 5-hydroxylase 1 | | Authors: | Stein, A.J, Goldberg, D.R, De Lombaert, S, Holt, M.C. | | Deposit date: | 2016-07-26 | | Release date: | 2017-01-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Optimization of spirocyclic proline tryptophan hydroxylase-1 inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
7CBT
 
 | | The crystal structure of SARS-CoV-2 main protease in complex with GC376 | | Descriptor: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase | | Authors: | Shi, Y, Peng, G. | | Deposit date: | 2020-06-13 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.346 Å) | | Cite: | The preclinical inhibitor GS441524 in combination with GC376 efficaciously inhibited the proliferation of SARS-CoV-2 in the mouse respiratory tract.
Emerg Microbes Infect, 10, 2021
|
|
5L2H
 
 | | Crystal Structure of W26A mutant of anti-EGFR Centyrin P54AR4-83v2 | | Descriptor: | Centyrin, GLYCEROL | | Authors: | Cardoso, R.M.F, Goldberg, S.D, O Neil, K.T, Gilliland, G.L. | | Deposit date: | 2016-08-01 | | Release date: | 2016-11-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8013 Å) | | Cite: | Engineering a targeted delivery platform using Centyrins.
Protein Eng. Des. Sel., 29, 2016
|
|
1NPQ
 
 | | structure of a rhodamine-labeled N-domain Troponin C mutant (Ca2+ saturated) in complex with skeletal Troponin I 115-131 | | Descriptor: | CALCIUM ION, Troponin C, Troponin I | | Authors: | Mercier, P, Ferguson, R.E, Irving, M, Corrie, J.E.T, Trentham, D.R, Sykes, B.D. | | Deposit date: | 2003-01-18 | | Release date: | 2003-04-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of a Bifunctional Rhodamine Labeled N-Domain of Troponin C Complexed with the Regulatory
"Switch" Peptide from Troponin I: Implications for in Situ Fluorescence Studies in Muscle Fibers
Biochemistry, 42, 2003
|
|
1BUA
 
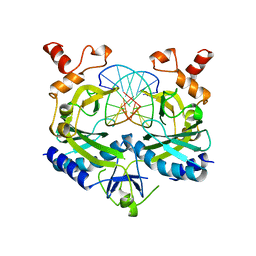 | |
1BSU
 
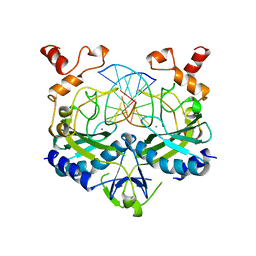 | |
2V3L
 
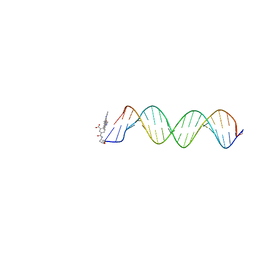 | | Orientational and dynamical heterogeneity of Rhodamine 6G terminally attached to a DNA helix | | Descriptor: | 5'-D(*CP*AP*AP*AP*GP*CP*GP*CP*CP*AP *TP*TP*CP*GP*CP*CP*AP*TP*TP*C)-3', 5'-D(*GP*AP*AP*TP*GP*GP*CP*GP*AP*AP *TP*GP*GP*CP*GP*CP*TP*TP*TP*G)-3', RHODAMINE 6G | | Authors: | Neubauer, H, Gaiko, N, Berger, S, Schaffer, J, Eggeling, C, Tuma, J, Verdier, L, Seidel, C.A.M, Griesinger, C, Volkmer, A. | | Deposit date: | 2007-06-18 | | Release date: | 2007-10-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Orientational and Dynamical Heterogeneity of Rhodamine 6G Terminally Attached to a DNA Helix Revealed by NMR and Single-Molecule Fluorescence Spectroscopy.
J.Am.Chem.Soc., 129, 2007
|
|
4U1F
 
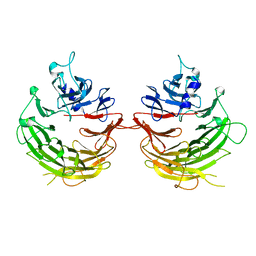 | |
2JR4
 
 | | NMR Solution Structure of the Anticodon of E.coli TRNA-VAL3 With no Modifications | | Descriptor: | 5'-R(*CP*CP*UP*CP*CP*CP*UP*UP*AP*CP*AP*AP*GP*GP*AP*GP*G)-3' | | Authors: | Vendeix, F.A.P, Dziergowska, A, Gustilo, E.M, Graham, W.D, Sproat, B, Malkiewicz, A, Agris, P.F. | | Deposit date: | 2007-06-20 | | Release date: | 2007-07-24 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Anticodon domain modifications contribute order to tRNA for ribosome-mediated codon binding.
Biochemistry, 47, 2008
|
|
2JRG
 
 | | NMR solution structure of the anticodon of E. coli TRNA-VAL3 with 2 modifications (cmo5U34 M6A37) | | Descriptor: | 5'-R(*CP*CP*UP*CP*CP*CP*UP*(CM0)P*AP*CP*(6MZ)P*AP*GP*GP*AP*GP*G)-3' | | Authors: | Vendeix, F.A, Dziergowska, A, Gustilo, E.M, Graham, W.D, Sproat, B, Malkiewicz, A, Agris, P.F. | | Deposit date: | 2007-06-26 | | Release date: | 2007-07-24 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Anticodon domain modifications contribute order to tRNA for ribosome-mediated codon binding.
Biochemistry, 47, 2008
|
|
1MD3
 
 | | A folding mutant of human class pi glutathione transferase, created by mutating glycine 146 of the wild-type protein to alanine | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE, pi glutathione transferase | | Authors: | Kong, G.K.-W, Dragani, B, Aceto, A, Cocco, R, Mannervik, B, Stenberg, G, McKinstry, W.J, Polekhina, G, Parker, M.W. | | Deposit date: | 2002-08-06 | | Release date: | 2002-08-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Contribution of Glycine 146 to a Conserved Folding Module Affecting Stability and Refolding of Human Glutathione Transferase P1-1
J.Biol.Chem., 278, 2003
|
|
1L9L
 
 | | GRANULYSIN FROM HUMAN CYTOLYTIC T LYMPHOCYTES | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, ETHANOL, Granulysin, ... | | Authors: | Anderson, D.H, Sawaya, M.R, Cascio, D, Ernst, W, Krensky, A, Modlin, R, Eisenberg, D. | | Deposit date: | 2002-03-25 | | Release date: | 2002-11-06 | | Last modified: | 2017-09-13 | | Method: | X-RAY DIFFRACTION (0.92 Å) | | Cite: | Granulysin Crystal Structure and a Structure-Derived Lytic Mechanism
J.Mol.Biol., 325, 2002
|
|
1JVM
 
 | |
1MD4
 
 | | A folding mutant of human class pi glutathione transferase, created by mutating glycine 146 of the wild-type protein to valine | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE, pi glutathione transferase | | Authors: | Kong, G.K.-W, Dragani, B, Aceto, A, Cocco, R, Mannervik, B, Stenberg, G, McKinstry, W.J, Polekhina, G, Parker, M.W. | | Deposit date: | 2002-08-06 | | Release date: | 2002-08-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Contribution of Glycine 146 to a Conserved Folding Module Affecting Stability and Refolding of Human Glutathione Transferase P1-1
J.Biol.Chem., 278, 2003
|
|
3STV
 
 | |
3STW
 
 | |
6N13
 
 | | UbcH7-Ub Complex with R0RBR Parkin and phosphoubiquitin | | Descriptor: | E3 ubiquitin-protein ligase parkin, Ubiquitin-conjugating enzyme E2 L3, ZINC ION, ... | | Authors: | Condos, T.E.C, Dunkerley, K.M, Freeman, E.A, Barber, K.R, Aguirre, J.D, Chaugule, V.K, Xiao, Y, Konermann, L, Walden, H, Shaw, G.S. | | Deposit date: | 2018-11-08 | | Release date: | 2018-11-28 | | Last modified: | 2020-01-08 | | Method: | SOLUTION NMR | | Cite: | Synergistic recruitment of UbcH7~Ub and phosphorylated Ubl domain triggers parkin activation.
EMBO J., 37, 2018
|
|
6G04
 
 | | NMR Solution Structure of Yeast TSR2(1-152) in Complex with S26A(100-119) | | Descriptor: | 40S ribosomal protein S26-A, Pre-rRNA-processing protein TSR2 | | Authors: | Michel, E, Schuetz, S, Damberger, F.F, Allain, F.H.-T, Panse, V.G. | | Deposit date: | 2018-03-16 | | Release date: | 2018-09-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Molecular basis for disassembly of an importin:ribosomal protein complex by the escortin Tsr2.
Nat Commun, 9, 2018
|
|
6VJV
 
 | | Crystal structure of the Prochlorococcus phage (myovirus P-SSM2) ferredoxin at 1.6 Angstroms | | Descriptor: | ACETATE ION, FE2/S2 (INORGANIC) CLUSTER, Ferredoxin, ... | | Authors: | Olmos Jr, J.L, Campbell, I.J, Miller, M.D, Xu, W, Kahanda, D, Atkinson, J.T, Sparks, N, Bennett, G.N, Silberg, J.J, Phillips Jr, G.N. | | Deposit date: | 2020-01-17 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Prochlorococcusphage ferredoxin: structural characterization and electron transfer to cyanobacterial sulfite reductases.
J.Biol.Chem., 295, 2020
|
|
2CH5
 
 | | Crystal structure of human N-acetylglucosamine kinase in complex with N-acetylglucosamine | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Weihofen, W.A, Berger, M, Chen, H, Saenger, W, Hinderlich, S. | | Deposit date: | 2006-03-13 | | Release date: | 2006-09-18 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of Human N-Acetylglucosamine Kinase in Two Complexes with N-Acetylglucosamine and with Adp/Glucose: Insights Into Substrate Specificity and Regulation.
J.Mol.Biol., 364, 2006
|
|
3STX
 
 | |
6G03
 
 | | NMR Solution Structure of yeast TSR2(1-152) | | Descriptor: | Pre-rRNA-processing protein TSR2 | | Authors: | Michel, E, Schuetz, S, Damberger, F.F, Allain, F.H.-T, Panse, V.G. | | Deposit date: | 2018-03-16 | | Release date: | 2018-09-19 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Molecular basis for disassembly of an importin:ribosomal protein complex by the escortin Tsr2.
Nat Commun, 9, 2018
|
|
2CH6
 
 | | Crystal structure of human N-acetylglucosamine kinase in complex with ADP and glucose | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, N-ACETYL-D-GLUCOSAMINE KINASE, alpha-D-glucopyranose | | Authors: | Weihofen, W.A, Berger, M, Chen, H, Saenger, W, Hinderlich, S. | | Deposit date: | 2006-03-13 | | Release date: | 2006-09-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Structures of Human N-Acetylglucosamine Kinase in Two Complexes with N-Acetylglucosamine and with Adp/Glucose: Insights Into Substrate Specificity and Regulation.
J.Mol.Biol., 364, 2006
|
|
2XND
 
 | | Crystal structure of bovine F1-c8 sub-complex of ATP Synthase | | Descriptor: | ATP SYNTHASE LIPID-BINDING PROTEIN, MITOCHONDRIAL, ATP SYNTHASE SUBUNIT ALPHA, ... | | Authors: | Watt, I.N, Montgomery, M.G, Runswick, M.J, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 2010-08-02 | | Release date: | 2010-09-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Bioenergetic Cost of Making an Adenosine Triphosphate Molecule in Animal Mitochondria.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
8VFU
 
 | |
